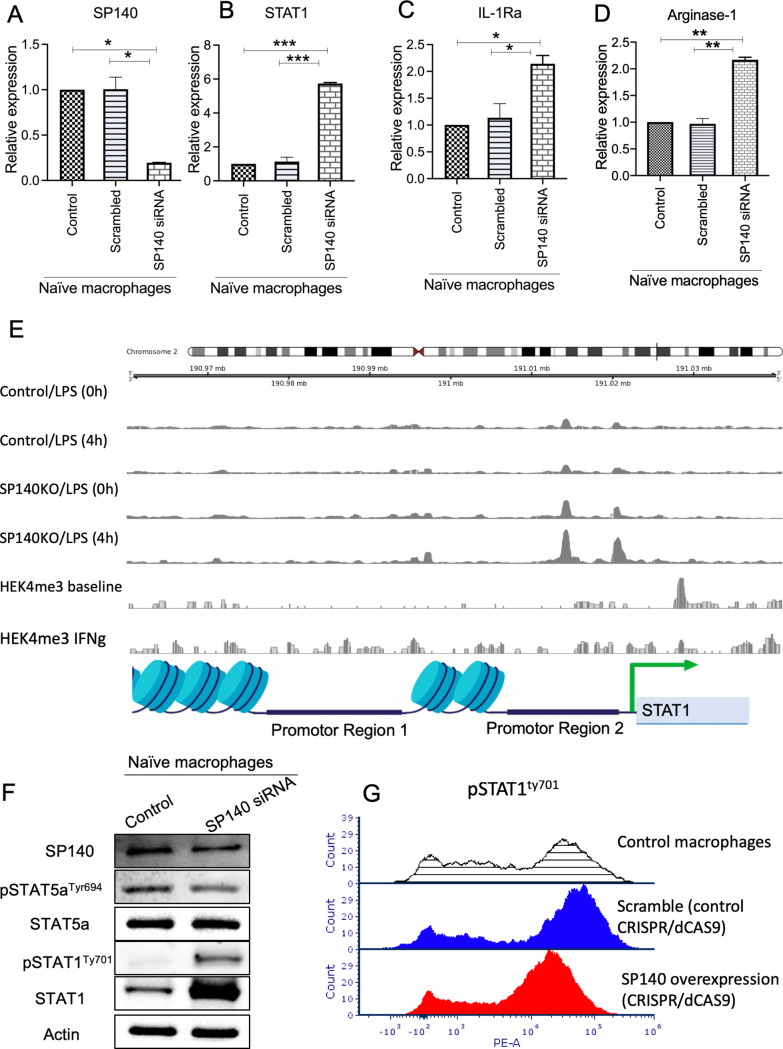
Figure 5.
SP140 negatively regulates STAT1 transcription and phosphorylation in macrophages. (A–D) SP140 was downregulated using siRNA in naïve (undifferentiated) macrophages, and cells were collected for RNA expression analysis by qPCR for SP140, STAT1, IL-1Ra and arginase-1, after 24 hours. 18S was used as an endogenous normalizer. The delta–delta Ct method was used to identify the relative gene expression. Data are presented as mean and SD of fold change compared with the control. (E) ATAC-seq read coverage (average count over an arbitrary sliding window) of SP140 KO and non-KO macrophages treated with LPS for 4 hours (along with their respective 0-hour controls) and average H3K27me3 ChIP-seq coverage for IFN-γ stimulated and unstimulated macrophages in the neighborhood of the STAT1 gene. (F) SP140 was downregulated using siRNA in naïve (undifferentiated) macrophages, and cells were collected for protein isolation and western blot 48 hours post transfection. (I) Effect of SP140 overexpressed on Levels of pSTAT1Tyr701 was quantified by flow cytometry in controls (shaded line indicates control macrophages; blue line indicates scramble control (CRISPR/dCAS9 control), and red line indicates test sample (SP140 overexpression, CRISPR/dCAS9)) after 48 hours. *P<0.05, **P<0.01, ***P<0.001. ATAC-seq, assay of transposase accessible chromatin sequencing; CHiP, chromatin immunoprecipitation; IFN-γ, interferon gamma; IL, interleukin; qPCR, quantitative PCR; siRNA, small interfering RNA; SP140, speckled protein 140.