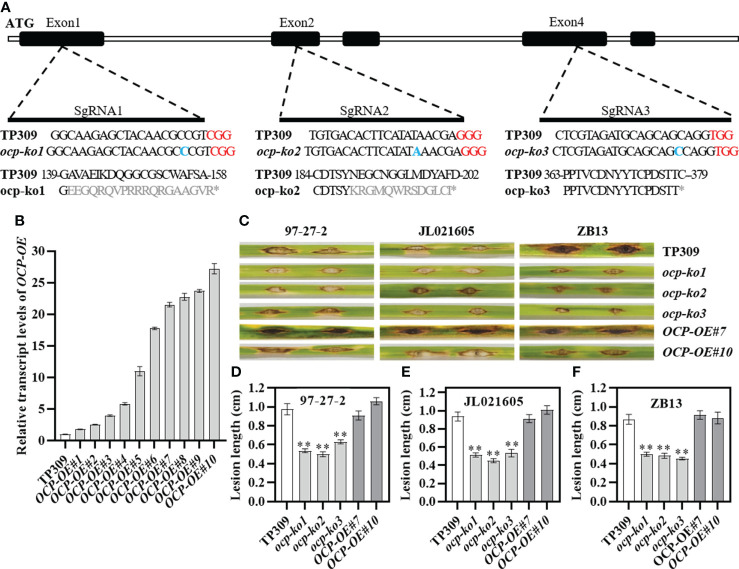
Figure 2.
Mutant types and blast resistance identification of OCP. (A) Three kinds of allelic variations induced by CRISPR/Cas9 in different regions. CDS are shown in the black boxes. Untranslated regions (UTR) and introns correspond to the white sections. The sgRNA1, sgRNA2, and sgRNA3 were located in Exon 1, Exon 2, and Exon 4, respectively, corresponding to three domains of OCP, inhibitor I29, pept_C1, and GRAN. Red bases indicate the protospacer adjacent motif (PAM) recognition sites. Blue bases show the insert location. In gray are the altered amino acid residues due to mutation. The * indicates the terminate codon. (B) Relative transcript levels of 10 overexpression lines (OCP-OE#1 to OCP-OE#10) in T1 of OCP. (C) Punch inoculation of wild-type rice TP309 and OCP mutant lines with the blast isolate 97-27-2, JL021605, and ZB13. This experiment was repeated twice. (D–F) Lesion lengths (mean ± SEM, n ≥ 8) of the tested lines according to the results in (C). Asterisks indicate statistical significance compared with TP309 (** P ≤ 0.01, t test).