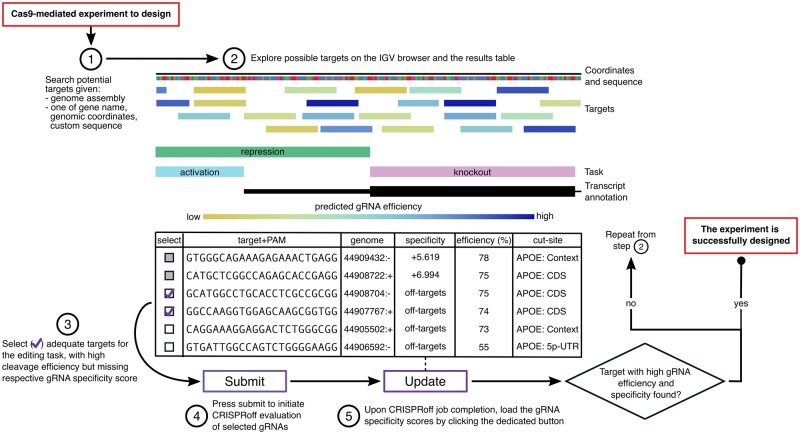
Fig. 1.
Design of gRNAs for Cas9 experiments with CRISPRon/off. Sequence of steps to identify targets that can be edited by Cas9 with high efficiency and with minimum off-target potential. The genomic sequence reported in the table rows is that of the DNA target (5′–3′ strand) which has the same sequence as the gRNA (which binds to the complimentary DNA) and the PAM. The ‘efficiency’ is the predicted indel frequency. The ‘specificity’ is the log10-scaled probability of binding at the on-target site compared to binding anywhere in the genome. The ‘genome’ coordinate is the start position of the target followed by the strand; all targets are in the same region and the same chromosome, which is not shown. IGV, integrative genomic viewer; CDS, coding sequence; 5p-UTR, 5′ untranslated region