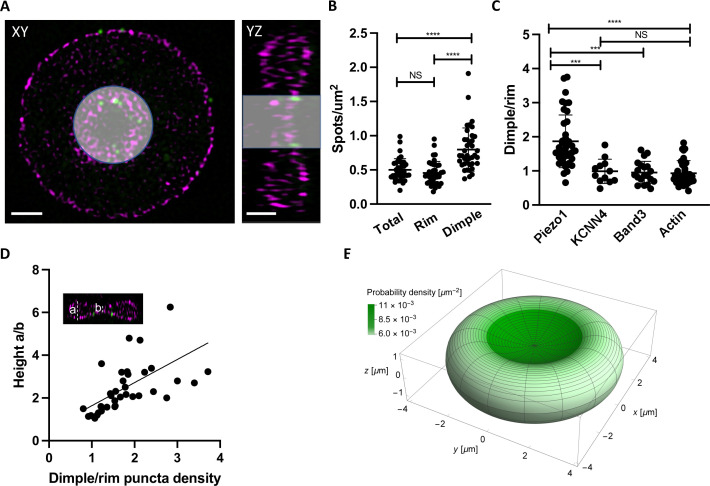
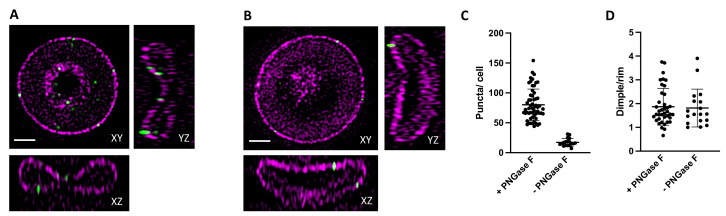
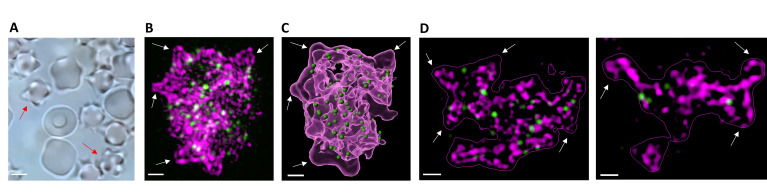
Figure 3. Piezo1 localization is enriched in the curved dimple regions of the red blood cell .
(A) Two dimensional slices in XY and YZ views from SIM images illustrating volume segmentation of RBCs and Piezo1 puncta distribution after labeling with an antibody against Piezo1-HA (green) and rhodamine–phalloidin for F-actin (magenta). Cells were segmented and masked in Imaris as shown. Scale bar, 1 μm. (B) Distribution of Piezo1 puncta in RBCs. The RBC dimple has a higher density of Piezo1 puncta than whole RBCs (Total) (p = <0.0001) and the rim region (p = <0.0001) by the one-way analysis of variance (ANOVA) test with Tukeys post hoc analysis, as indicated on the graph by asterisks where *** indicates p<0.001 and **** indicates p<0.0001. N = 39 cells (C) The ratio of Piezo1 spot density (1.87 ± 0.76, N = 39 cells) in the dimple and rim region of RBCs compared to that of KCNN4 (0.99 ± 0.36, N = 12 cells), Band3 (0.95 ± 0.33, N = 20 cells), and Actin (0.93 ± 0.36, N = 31 cells). The dimple over rim ratio is higher for Piezo1 than KCNN4 (p = 0.0003) and Band3 (p = 0.0003) and Actin (p < 0.0001). (D) Scatterplot of ratio of Piezo1 puncta density in dimple over rim plotted against RBC biconcavity, measured as height ratio a/b (see inset). N = 37 cells. Data are fit by linear regression with equation y = 1.08x + 0.55, R2 = 0.34 and p = 0.0001. (E) Probability per unit area for finding a Piezo1 channel at a given RBC membrane location, calculated from a physical model of Piezo1 curvature coupling (see methods) and plotted over the RBC membrane surface. The RBC shape corresponds to Beck’s model of for RBC discocytes (Beck, 1978).