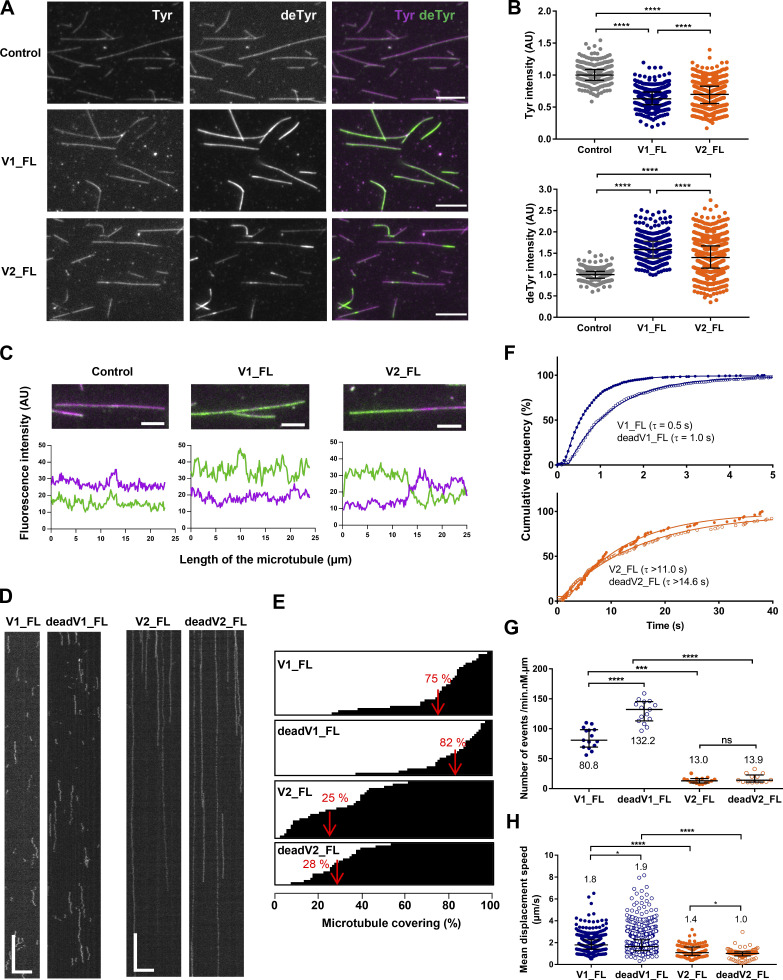
Figure 1.
VASH1–SVBP and VASH2–SVBP complexes exhibit dissimilar microtubule detyrosination activities and very different binding behaviors on microtubules. (A−C) Comparison of the activity of sfGFP-tagged VASH1–SVBP (V1_FL) and VASH2–SVBP (V2_FL) enzyme complexes (50 pM) on Taxol-stabilized Tyr-MTs measured by immunofluorescence in BRB40 supplemented with 50 mM KCl. (A) Representative images of tyrosinated (Tyr, magenta) and detyrosinated (deTyr, green) α-tubulin pools of microtubules after 30 min incubation in the absence (control) or presence of the indicated enzyme complexes. Scale bar, 10 µm. (B) Analysis of tyrosinated- and detyrosinated-tubulin signal intensity. Each point represents a microtubule (at least 300 microtubules were analyzed). Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test, ****P < 0.0001. (C) Graphs of fluorescence intensity variations of tyrosinated (magenta) and detyrosinated (green) tubulin on selected microtubules in the absence and presence of enzyme. Scale bar, 5 µM. (D−H) TIRF microscopy study of single molecules of sfGFP-tagged VASH1–SVBP (V1_FL and deadV1_FL) and VASH2–SVBP (V2_FL and deadV2_FL) bound to microtubules in the same experimental conditions as in A–C. (D) Representative kymographs. Scale bars: horizontal, 5 µm; vertical, 5 s. Examples of TIRF movies for active VASH1–SVBP and VASH2–SVBP from which kymographs were extracted are presented in supplemental data (Videos 1 and 2). (E−H) Analysis of the binding characteristics. Results for VASH1–SVBP are in blue and for VASH2–SVBP in orange, with plain circles for active enzymes (V1_FL and V2_FL) and empty circles for their catalytically dead versions (deadV1_FL and deadV2_FL). (E) Representation of the microtubule surface covered with VASH–SVBP molecules (white) or not covered (black) during the 45 s of a TIRF movie. Each horizontal line represents a microtubule (at least 19 microtubules were analyzed). The red value corresponds to the mean covering (in %). (F) Cumulative frequency of the residence times measured in TIRF movies taken during the 30 min following addition of enzyme complexes to microtubules. The mean residence time (τ) is obtained by fitting the curve with a mono-exponential function. (G) Analysis of binding frequency. Each point represents a microtubule. (H) Analysis of diffusion. Each point represents a single molecule, the molecules moving on at least 15 microtubules were analyzed. Data are represented as the median with the interquartile range. Statistical significance was determined using Kruskal-Wallis test. *P < 0.01, ***P < 0.005, ****P < 0.0001. Binding characteristics are also summarized in Table 1.