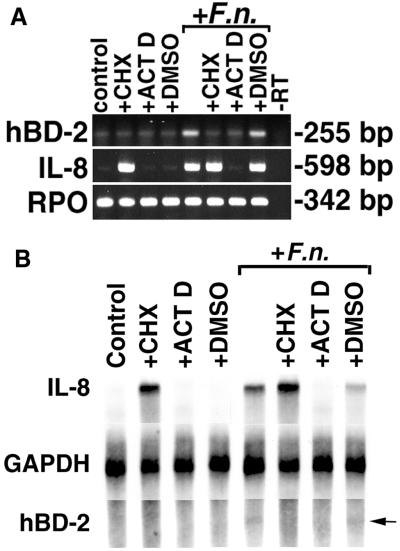
FIG. 9.
Complete inhibition of hBD-2 induction by pretreatment of HGE with cycloheximide and actinomycin D. (A) RT-PCR analysis. HGE were pretreated with 10 μg of cycloheximide/ml (+CHX), 1 μg of actinomycin D/ml (+ACT D), or DMSO solvent (+DMSO) for these two reagents for 1 h, and some samples were then challenged with 10 μg of F. nucleatum cell wall extract/ml (+F.n.) for 6 h. The 6-h time point was chosen because overnight incubation of HGE with cycloheximide and actinomycin D was toxic. At 6 h, there was no significant difference in viability between control and treated cells as assessed by staining with trypan blue. Furthermore, the yields of total RNA from the samples were compared and showed no difference. Total RNA was extracted and analyzed by RT-PCR. Note that in this experiment, PCR for hBD-2 was performed for 28 cycles, which resulted in an increased background of hBD-2 expression in the unstimulated (control) sample and the samples treated with CHX or ACT D in the presence or absence of F. nucleatum cell wall. The data shown are representative of three independent experiments. −RT, minus-RT control. (B) RPA analysis. Total RNA (20 μg) from the samples in panel A was analyzed by RPA as described in Materials and Methods. GAPDH hybridization is shown as a normalization control. Note that hBD-2 mRNA induction is inhibited with both CHX and ACT D, while IL-8 mRNA expression is inhibited only with ACT D. Also note the superinduction of IL-8 mRNA in the sample treated with both CHX and F. nucleatum cell wall and that the degree of hBD-2 expression was lower than that in Fig. 3A due to a shorter time of stimulation (6 h). The arrow indicates a protected fragment of hBD-2 in the samples treated with F. nucleatum and F. nucleatum cell wall plus DMSO.