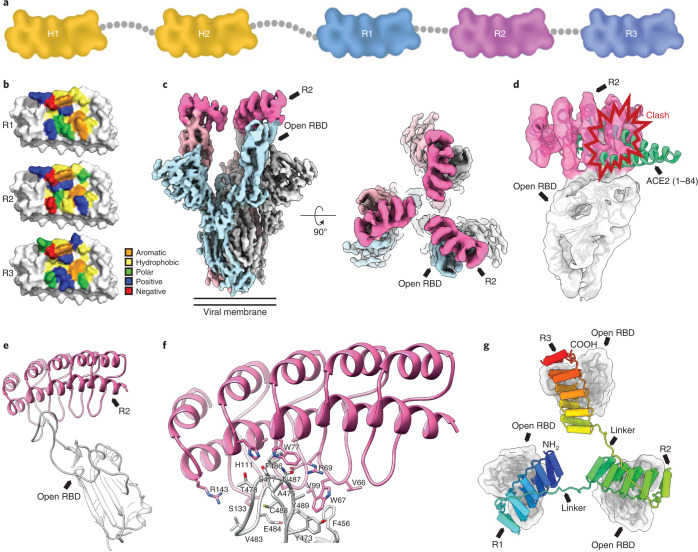
Fig. 1. Structural modeling of ensovibep.
a, Schematic overview of the ensovibep construct. Protein linkers are depicted as gray dashed lines, and the half-life-extending HSA-binding monovalent DARPins (H1 and H2) are colored yellow. b, Surface representations of the three monovalent DARPin molecules binding to the RBD, with the amino acid residues in the paratope colored according to their biophysical characteristics as indicated. c, cryo-EM density for the SARS-CoV-2 spike ectodomain in complex with the RBD-targeting monovalent DARPin R2, shown as two orthogonal views. The DARPin density is colored magenta, and the three spike protomers are colored light blue, gray and pale pink. d, Zoomed-in view of an RBD bound to DARPin R2 with the cryo-EM density shown semi-transparent. The atomic coordinates for the fitted open RBD (PDB ID: 6XCN) and the DARPin model are overlaid. The atomic coordinates for residues 1–84 of the RBD-bound ACE2 (PDB ID: 6M0J), colored green, are superimposed. e, Pseudo-atomic model of the monovalent DARPin R2 in complex with the RBD, colored pink and gray, respectively. f, Zoomed-in view of the interface between monovalent DARPin R2 and RBD. g, Proposed model of the three covalently linked RBD-targeting monovalent DARPin molecules of ensovibep bound to the trimeric spike protein RBD domains. The three DARPin domains are shown in a rainbow color scheme from the N terminus (blue) to the C terminus (red).