Summary
The protocol is designed to investigate the influence of an anti-cleavage site intrabody in modulating the output of LV(CoV-2 S), a lentivirus-based pseudovirus expressing CoV-2 S protein using HEK293T cells. We clone the single-domain antibody sequence into a lentiviral vector (pLenti-GFP) for intracellular expression and assess not only the viral biogenesis but also the fate of the CoV-2 S protein in such cells.
For complete details on the use and execution of this protocol, please refer to Singh et al. (2022).1
Subject areas: Cell Biology, Immunology, Molecular Biology
Graphical abstract

Highlights
-
•
Clone an anti-CSP sdAb into pLenti-GFP vector for intracellular expression
-
•
Generate LV(CoV-2 S) pseudovirus from anti-CSP IB-expressing HEK293T cells
-
•
Quantify pseudovirus made from anti-CSP IB-expressing cells by infecting Vero E6 cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The protocol is designed to investigate the influence of an anti-cleavage site intrabody in modulating the output of LV(CoV-2 S), a lentivirus-based pseudovirus expressing CoV-2 S protein using HEK293T cells. We clone the single-domain antibody sequence into a lentiviral vector (pLenti-GFP) for intracellular expression and assess not only the viral biogenesis but also the fate of the CoV-2 S protein in such cells.
Before you begin
This protocol provides detailed methodology used in the published article; Robust anti-SARS-CoV-2 single domain antibodies cross neutralize multiple viruses.1 The protocol describes a strategy for the intracellular expression of sdAbs targeting the polybasic cleavage site (anti-CSP sdAb) of the viral entry mediator, SARS-CoV-2 S protein. We also analyze the intracellular fate of the protein in the anti-CSP intrabody (IB)-expressing cells.
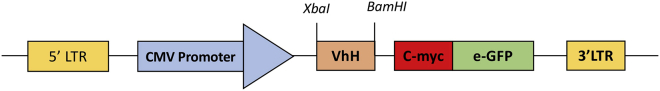
For generating an anti-CSP IB construct, the sequence of the selected sdAb specific to peptide (NSPRRAR/SVAS) which encompasses the polybasic cleavage site (CSP) of SARS-CoV-2 S protein was cloned into a pLenti-GFP vector (a lentivirus-based mammalian expression vector) downstream to CMV promoter.2 The cloning scheme used is: 5′LTR-CMV promoter-VHH - c-Myc-tag - e-GFP -3′-LTR. The anti-CSP sdAb sequence was amplified using the forward primer, VHH-FR1-pLenti (containing XbaI restriction site), and reverse primer, FR4-c-Myc pLenti, that encoded c-Myc tag at 3′ end of VHH with a BamHI site) (Figure 1).
Figure1.
Cloning strategy of anti-CSP intrabody into pLenti-GFP vector
To produce anti-CSP sdAb as intrabody, the sdAb sequences were cloned in frame with a c-Myc and/or GFP to generate a fusion product. The cloning strategy for making the anti-CSP IB construct, 5′LTR-CMV promoter-VHH - c-Myc-tag - e-GFP -3′-LTR. XbaI and Bam HI restriction enzyme were used for cloning anti-CSP sdAb into the pLenti-GFP vector.
To generate the lentivirus-based pseudoviruses expressing surface SARS-CoV-2 S protein, HEK293T cells were transfected with plasmids such as a reporter construct, pCMVR8.74, SARS-CoV-2 S construct, tat and rev as well as pMD2.G.3 The plasmids required for producing LV(CoV-2 S) pseudoviruses were prepared and the purity was confirmed before beginning with the experiments.
Institutional permission
Requisite approval from the institutional biosafety committee were obtained and the experiments were conducted strictly in accordance with the approved protocols.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-c-Myc tag antibody (1:2000) | Santa Cruz Biotech (SCBT) | Cat#9E10 |
| Streptavidin APC (1:200) | BioLegend | Cat#405207 |
| Mouse anti-6×(HIS) tag (1:5000 for WB, 1:100 in confocal) | Invitrogen | Cat#4A12E6 |
| Alkaline phosphatase-conjugated goat anti-mouse IgG (1:10000 for WB) | Sigma | Cat#A3562 |
| Anti-Ubiquitin antibody (1:2500) | SCBT | Cat#Sc8017 |
| Anti-GAPDH antibody (1:1500) | Invitrogen | TG267131 |
| Golgin (rabbit) (1:100) | Sigma | PA5-52841 |
| Calnexin (rabbit) (1:100) | Cell Signal Technology (CST), Beverly, Massachusetts, USA | 52533S |
| Goat anti-mouse Alexa Fluor 647 (1:100) | Thermo Fisher Scientific | A21245 |
| Goat anti-Rabbit Alexa fluor 568 (1:100) | Thermo Fisher Scientific | A11036 |
| Hoechst stain (10 μg/mL) | Thermo Fisher Scientific | 62249 |
| Streptavidin HRP (1:500) | Life Technology | SNN1004 |
| Anti-Flag mouse antibody (1:2500) | Sigma | F1804-50UG |
| Anti-CSP sdAb (10 μg/mL) | In-house generated | Ref.1 |
| Chemicals, peptides, and recombinant proteins | ||
| Tunicamycin | Sigma | T7765-5MG |
| PVDF membrane | Bio-Rad | Cat#1620177 |
| Trypsin | HiMedia | 59427C-500mL |
| Bovine serum albumin | HiMedia | GRM105-100G |
| Ni- NTA beads | G Biosciences | Cat#786940 |
| TMB substrate for ELISA | Becton, Dickinson and Company (BD) | Cat#555214 |
| FemtoLUCENT plus AP kit | G-Biosciences | Cat#786-10AP |
| PEI branched | Sigma | 408727-100mL |
| BamHI | New England Biolab (NEB) | R3136S |
| XbaI | NEB | R0145S |
| NcoI | NEB | R3193S |
| NotI | NEB | R3189S |
| MG132 | Sigma | 474787 |
| Ni-NTA probes | ForteBio | 18-5101 |
| Clarity Western ECL substrate | Bio-Rad | Cat#170-5060 |
| pNPP substrate | Sigma | 71768-25G |
| DMEM | Gibco | 10566-016-500mL |
| RPMI | Gibco | A10491-01 500mL |
| Fetal bovine serum | Gibco | 10270106- 500 mL |
| NSPRRARSVAS (cleavage site peptide) | S-Biochem | Used in this study |
| Recombinant DNA | ||
| WT SARS-CoV-2 S plasmid | Dr. Jason McLellan (University of Texas)4 | https://doi.org/10.1126/science.abb2507 |
| pCMVR8.74 | Dr. Indranil Banerjee (IISER Mohali) | #22036 |
| pMD2.G | Dr. Indranil Banerjee | #12259 |
| pCMV14-3×-FLAG-SARS-CoV-2 S | Addgene | #145780 |
| pLenti-GFP | Addgene | #17448 |
| Tat 1b and Rev 1b | SARS-Related Coronavirus 2, Wuhan-Hu-1 spike-pseudotyped lentiviral kit | NR-52948 |
| pYBNT (modified pet 22b) Anti-CSP-IB clone |
Novagen In-house generated | pET22b+ Used in this study (Described earlier)5 |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | CRL-3216™ |
| Vero E6 cells | Dr. Rajesh Ringe lab (IMTECH, CHANDIGARH) | NA |
| Vero cells | ATCC | CCL-81 |
| Experimental models: Organisms/strains | ||
| DH5 alpha-competent cells | Ref.5 | Ref.5 |
| Origami competent cells | Ref.5 | Ref.5 |
| Stbl3 competent cells | Thermo Fisher Scientific | Cat#C7381201 |
| Software and algorithms | ||
| FlowJo software | BD Biosciences | https://www.flowjo.com/solutions/flowjo/downloads |
| Image J | National Institute of Health (NIH), MD | https://imagej.nih.gov/ij/download.html |
| Immune Epitope Data Base | National Institute of Allergy and Infectious Diseases (NIAID), MD | http://tools.iedb.org/main/bcell/ |
| BLItz software | ForteBio | http://www.blitzmenow.com |
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Flow cytometer | Accuri-C6 | BD |
| Fluorescence microscope | Nickon | Nikon eclipse Ti |
| Confocal microscope | Leica | SP8 upright |
| ELISA plate reader | Biotech | Eon |
| CO2 incubator | Eppendorf | Galaxy 170 R |
| Microbial incubator | Labquick | Talboys |
| Biosafety cabinet | Labconco | Logic+ |
| Centrifuge | Eppendorf | 5810R |
| Rotospin | Tarsons | Tarson |
| 100-mm petri dishes | CLS430167-500EA | Merck |
| 96 well-flat bottom plates | 3799 | Corning |
| Glass slides | Frosted Micro-slides | Blue star |
| Coverslips | Microscopic cover glass | Blue star |
| X-ray film | Edusoft Healthcare | E-Ray |
| FP VHH FR1 pLenti: 5′GCATT CTAGAGGCACCCCGGGATG GCCGATGTTCAACTGCAGGAG3′ |
GCC Biotech India | Ref.1 |
| FR4 C-Myc pLenti: 5′GCTAGG ATCCTGCAGATCCTCTTCAGAG ATGAGTTTCTGCTCTGTGGAGA CGGTGACCTG3′ |
GCC Biotech India | Ref.1 |
| MKS-9 (VHH FP): 5′GTTGTGT GGAATTGTGAGCG3′ |
GCC Biotech India | Ref.5 |
| MKS-22 (VHH RP): 5′GAAATGCGG CCGCTGTGGAGACGGTGACCTG3′ |
GCC Biotech India | Ref.5 |
| T7-FP: 5′TAATACGACT CACTATAGGGGAATTGTG-3′ |
GCC Biotech India | Ref.5 |
| T7-RP: 5′GCTAGTTATTGCT CAGCGGTGGCAGCAGC3′ |
GCC Biotech India | Ref.5 |
Materials and equipment
Recipes and reagent for preparing buffer and media
Reagents
| 1×-phosphate buffer saline (PBS) | Final conc. | Amount |
|---|---|---|
| Sodium Chloride | 137 mM | 8 g |
| Potassium Chloride | 2.7 mM | 0.201 g |
| Sodium di-hydrogen phosphate | 10 mM | 1.429 g |
| Potassium di-hydrogen phosphate | 2 mM | 0.26 g |
| Milli-Q water | Ad | 1 Lt |
Filter through a 0.2 μm sterile filter and store at 25°C.
| 1×-PBST | Final conc. | Amount |
|---|---|---|
| Sodium Chloride | 137 mM | 8 g |
| Potassium Chloride | 2.7 mM | 0.201 g |
| Sodium di-hydrogen phosphate | 10 mM | 1.429 g |
| Potassium di-hydrogen phosphate | 2 mM | 0.26 g |
| Tween-20 | 0.05% | 0.5 mL |
| Milli-Q water | Ad | 1Lt |
Filter through a 0.2 μm sterile filter and store at 25°C.
| 1×-PBS-EDTA (pH 7.4) | Final conc. | Amount |
|---|---|---|
| Sodium Chloride | 137 mM | 8 g |
| Potassium Chloride | 2.7 mM | 0.201 g |
| Sodium di-hydrogen phosphate | 10 mM | 1.429 g |
| Potassium di-hydrogen phosphate | 2 mM | 0.26 g |
| EDTA | 10 mM | 20 mL |
| Milli-Q water | Ad | 1 Lt |
Filter through a 0.2 μm sterile filter and store at 25°C.
Media
| 10% DMEM (pH 7.4) | Final conc. | Amount |
|---|---|---|
| Serum free DMEM | – | 445 mL |
| FBS | 10% | 50 mL |
| Pen strep (100×) | 1× | 5 mL |
| Total volume | – | 500 mL |
Filter through 0.2 μm sterile filter and store at 4°C for 1 month.
| LB medium for growing bacteria | Final conc. | Amount |
|---|---|---|
| LB powder | NA | 25 g |
| DD water | ad | 1 Lt |
Autoclaved for 15 min at 121°C and kept at 25°C.
| Plasmid isolation reagent | Final conc. | Amount |
|---|---|---|
| Buffer I (Resuspension buffer) | ||
| Glucose | 50 mM | 1.80 g |
| Tris-HCl (pH 8) | 25 mM | 605.7 mg |
| EDTA (pH 8) | 10 mM | 4 mL |
| Milli-Q water | Ad | 200 mL |
Autoclaved for 15 min at 121°C store at 4°C.
| Buffer II (lysis buffer) prepared freshly | Final conc. | Amount |
|---|---|---|
| Sodium Hydroxide | 10 N | 400 μL |
| Sodium dodecyl sulfate | 10% | 2 mL |
| Milli-Q water | Ad | 20 mL |
| Buffer III (neutralization buffer) | Final conc. | Amount |
|---|---|---|
| Potassium acetate | 5 M | 41.01 g |
| Glacial Acetic Acid | 11.5% | 11.5 mL |
| Milli-Q water | Ad | 100 mL |
Autoclaved for 15 min at 121°C and keep at 4°C.
| 4×lentivirus concentrator solution (pH 7.4) | Final conc. | Amount |
|---|---|---|
| Polyethylene glycol (PEG)-8000 | 40% (W/V) | 80 g |
| Sodium Chloride | 1.2 M | 14 g |
| 10×-PBS | 1× | 20 mL |
| Milli-Q water | Ad | 200 mL |
Autoclaved for 15 min at 121°C Filter through 0.2 μm and store at 25°C.
| Blocking solution for ELISA | ||
|---|---|---|
| Bovine serum albumin | 5% | 0.5 g |
| 1×-PBS+ 0.05% Tween-20 | Ad | 10 mL |
Prepare fresh before use.
| Blocking solution for western blotting | ||
|---|---|---|
| Skim milk | 5% | 0.5 g |
| 1×-PBS+ 0.05% Tween-20 | Ad | 10 mL |
Prepare fresh before use; filter through Whatman paper.
| Blocking solution confocal analysis | ||
| Bovine serum Albumin | 5% | 0.5 g |
| Triton X-100 | 0.3% | 30 μL |
| 1×-PBS | Ad | 10 mL |
| Antibody dilution solution for confocal experiments | ||
| Bovine serum Albumin | 1% | 0.1 g |
| Triton X-100 | 0.3% | 30 μL |
| 1×-PBS | Ad | 10 mL |
| C-100 buffer (pH 7.4) | ||
| HEPES | 20 mM | 953.2 mg |
| EDTA | 0.2 mM | 11.68 mg |
| MgCl2 | 1.5 mM | 28.56 mg |
| KCl | 100 mM | 1.49 g |
| Glycerol | 20% (V/V) | 40 mL |
| NP-40 | 0.02% (V/V) | 40 μL |
| Milli-Q water | Ad | 200 mL |
Step-by-step method details
Revival of HEK293T cells and Vero E6 cells
Timing: 3–10 days (for steps1–4)
This step will provide a brief procedure for revival of frozen cells.
-
1.
Take aliquots of HEK293T and Vero E6 cells stored at −80°C and thaw them immediately. Wash the cells once with 10% DMEM by centrifuging at 250 × g for 5 min at 25°C.
-
2.
Discard the supernatant and resuspend the cells pellet into 5 mL of 10% DMEM to seed the cells into a T25 cell culture flask. Keep the flasks in a CO2 incubator (37°C, 5% CO2, 95% relative humidity, RH).
-
3.
After 3rd-day of reviving the cells, replace the medium with freshly prepared complete DMEM (10% DMEM) and repeat the process until a monolayer is formed.
-
4.
On 7th day, split the cells into multiple tissue culture dishes of 100 mm for performing the experiments. On or after 10th day, when the cells are approximately 80% confluent, initiate the transfection experiments to generate pseudoviruses.
Transfecting HEK293T cells for the generation of LV(CoV-2 S) pseudoviruses
Timing: 3–4 days (for steps5–13)
We describe here the detailed methodology for generating the pseudoviruses using HEK293T cells and concentrating such particles by PEG-NaCl.
-
5.Production of pseudovirus.
-
a.Production of pseudoviruses using HEK293T cells not expressing anti-CSP IB: For generating replication-deficient LV(CoV-2 S) pseudoviruses, HEK293T cells cultured at ∼80% confluency in 100 mm dishes are co-transfected with five plasmids (stored at −20°C and thawed on ice) viz., 10 μg of pLenti-GFP (reporter plasmid with GFP), 9.8 μg of pCMVR8.74 (packaging vector), 6 μg of SARS-CoV-2 spike encoding plasmid, 6 μg each of Tat (trans-activator) and Rev plasmids (which promote the expression of virion proteins).
-
b.Production of pseudoviruses in anti-CSP IB-expressing HEK293T cells: Co-transfect HEK293T cells cultured at ∼80% confluency with 5 plasmids viz., 10 μg of anti-CSP IB construct in which the sequence of anti-CSP sdAb is cloned upstream of GFP using Xba1 and BamH1 sites (pLenti-GFP-anti-CSP sdAb GFP-reporter plasmid, 9.8 μg of pCMVR8.74, 6 μg of SARS-CoV-2 S construct, 6 μg each of Tat and Rev plasmids (trans activator and positive regulator of virion proteins expression, respectively). The cloning strategy is outlined in Figure 1.
-
a.
-
6.
To prepare the transfection mixture, aliquot the above-mentioned plasmids in a 1.5 mL microcentrifuge tube and add the plasmid mixture with the transfection agent, PEI in 1:3 ratios (37.8 μg of total 5 plasmids and 114 μL of PEI) in a final concentration of 1 μg/mL to make a final volume of 1 mL with serum-free DMEM. Vortex the mixture for 30 s and keep at 25°C for 15 min (Refer to Table 1).
-
7.
Carefully add the transfection mixture onto HEK293T cells, cultured in 100 mm plates and gently swirl the plates.
CRITICAL: Make sure that the incubation time for making nano-vesicles does not exceeds 20 min, as this will reduce the transfection efficiency.
-
8.
After 16 h of transfection, replace the medium with complete DMEM.
Note: This step will reduce PEI-induced detachment and death of the cells.
-
9.
After 72 h of transfection, collect the supernatant in separate falcon tubes from both the conditions (1, 2) and centrifuge the tubes at 100 × g to remove cell debris.
Note: Check for GFP expression using a fluorescent microscope to assess the percentage of transfected cells.
-
10.
Collect supernatant in separate falcons and add PEG-NaCl in 1:3 ratios, i.e., one part PEG-NaCl (can be stored at 25°C for a week) and 3 parts of the supernatant with pseudoviruses to concentrate the pseudovirus particles by gently rotating at 25rpm for 16 h at 4°C.
-
11.
Next day, centrifuge the concentrated pseudoviruses at 3,000 × g for 80 min at 4°C.
-
12.
Discard supernatant and resuspend the pellet containing pseudoviruses in serum-free DMEM.
Note: For the supernatant collected from at least 5 of the 100 mm plates, resuspend the pellet in 5 mL of serum-free DMEM.
-
13.
The collected supernatant can be used for further experiments.
Pause point: Concentrated pseudoviral particles can be stored at 4°C for a week. For long time storage keep the pseudoviruses at −80°C.
Table 1.
Recipe for preparing transfection mixture for transfection of one, 100 mm petri-dish
| Reagent | Final concentration | Amount |
|---|---|---|
| pLentiGFP/Anti-CSP IB construct | 10 μg/mL | 10 μg |
| pCMVR8.74 | 9.8 μg/mL | 9.8 μg |
| Spike construct | 6 μg/mL | 6 μg |
| Tat plasmid | 6 μg/mL | 6 μg |
| Rev plasmid | 6 μg/mL | 6 μg |
| polyethyleneimine (PEI) | 1 μg/mL | 114 μL |
| Serum Free DMEM | ad | 1 mL |
Assessing the infectivity of LV(CoV-2 S) pseudoviruses produced from the control cells and those expressing anti-CSP IB
Timing: 3–4 days (for steps14–20)
This section includes the detailed procedure for measuring the infectivity of the generated pseudoviruses in Vero E6 cells.
-
14.
Seed approximately 10,000 Vero E6 cells in 96 flat-bottom plates and keep the plate in a CO2 incubator (37°C, 5% CO2, 95% RH) for 16 h.
-
15.
Infect Vero E6 cells at 80% confluency with 200 μL/well of pseudoviruses from each of the above conditions.
-
16.
After 16 h of infection, replace the medium with complete DMEM and observe for GFP expression using a fluorescent microscope.
-
17.
After 72 h of infection, capture images from different wells along with those from the control wells in which only LV(BALD) particles are added.
-
18.
Collect the infected and control cells from all the wells using 1×-PBS-EDTA (can be stored at 25°C for a week) and then wash twice with 1×-PBS (can be stored at 25°C for a week) to remove clumps of dead cells.
CRITICAL: Incubate the cells at 37°C for 15 min in a CO2 incubator to increase the isolation efficiency.
-
19.
Acquire and analyze the infected cells using a flow cytometer. To measure the frequency of GFP-positive i.e., the infected cells in different conditions, insert a gate to mark GFP-negative cells by using the LV(BALD) infected cells as a negative control. During the acquisition, remove all the debris by setting up a threshold of 5,00,000.
-
20.
Collect data and transform the above files as FCS files (if samples are acquired using a BD C6 Accuri) for analyzing the above samples in FlowJo software (Schematic is shown in Figure 2 and the data is shown in Figure 5 of the original article Singh et al.1).
Note: While performing the analysis, first gating is done to include total live cells and then the GFP-positive cells are measured in different conditions.
Figure 2.
Schematic of production of LV(CoV-2 S) based pseudoviruses from transfected HEK293T cells (control and anti-CSP IB condition)
The transduction efficiency was measured using fluorescence microscopy and flow cytometry. Supernatant were also used for measuring the SARS-CoV-2 S protein assembled in the LV(CoV-2 S) pseudoviruses using ELISA.
Figure 5.
Flow cytometric analysis of the transfected HEK293T cells for differential expression of SARS-CoV-2 S protein
Transfected HEK293T cells were scrapped out and washed twice with 1×-PBS. One aliquot of the cells was stained for flow cytometric analysis to analyze the presence of surface SARS-CoV-2 S protein using anti-CSP-sdAb (biotinylated anti-CSP sdAb). Streptavidin-APC was used to visualize the expression of SARS-CoV-2 S protein. The cells in the other groups were fixed, permeabilized and stained for detecting the intracellular expression of SARS-CoV-2 S protein.
(A) Transfected HEK293T cells were checked for GFP expression and streptavidin-APC positive cells as control.
(B) Transfected HEK293T cells were stained for the presence of surface and intracellular SARS-CoV-2 S protein expression as indicated.
(C) Bar diagrams show cumulative result from experiments performed in (B).The levels of statistical significance were measured by two-way ANOVA, ∗∗∗p<0.001, ∗p<0.01 and ∗p<0.05.
Assessing the production of LV(CoV-2 S) pseudovirus particles produced from control and anti-CSP IB by ELISA
Timing: 1–2 days (for steps21–29)
This section describes the method to measure differences in the titers of pseudoviruses produced in the supernatant of transfected HEK293T cells in the presence or absence of anti-CSP IB using ELISA.
-
21.
Coat the wells of ELISA plates in triplicate with 100 μL each of the concentrated pseudovirus preparations (those obtained from control and the anti-CSP IB-expressing cells) and keep the plates at 4°C for 16 h.
Note: Include negative controls such as serum-free DMEM, LV(BALD) pseudovirus particles, and supernatant from the cultured Vero E6 cells.
-
22.
Next day, wash the wells four times with 200 μL of 0.05% PBST (store at 25°C, for maximum of 3 days). For every washing, gently rotate the plates for 3–4 min at 25°C.
-
23.
Add 200 μL of blocking buffer (5% BSA in PBST) in each well and incubate for 2 h at 25°C. Wash the plate four times with 0.05% PBST.
-
24.
Add biotinylated anti-CSP sdAb (final conc. 10 μg/mL) diluted in 1×-PBS to each of the wells and incubate for 2 h at 25°C.
-
25.
Wash the plates four times with 0.05% PBST.
-
26.
Add HRP-conjugated streptavidin (1:500 diluted in 0.05% PBST) for 45 min and incubate the plates at 25°C.
-
27.
Wash the plate five times with 0.05% PBST.
-
28.
Add 100 μL of TMB substrate (substrate reagent A: substrate reagent B in 1:1 ratio) to develop the reaction. Stop the reaction by adding 50 μL of ortho-phosphoric acid.
-
29.
Measure the OD values at a wavelength of 450 nm using an ELISA reader (Schematic is shown in Figures 2 and the data is shown in Figure S7D of the original article Singh et al.1).
Assessing the infectivity of LV(CoV-2 S) pseudovirus particles produced from the control and anti-CSP IB-expressing cells using confocal microscopy
Timing: 2–3 days (for steps30–44)
In this section, we show the interaction of SARS-CoV-2 S protein and intrabody within HEK293T cells using confocal microscopy.
-
30.
Treat the required number of coverslips with 1 N HCl solution and wash in Milli-Q water. Then autoclave the coverslips.
-
31.
Wash these coverslips with autoclaved Milli-Q water for 10 min, followed by sterilization with UV light for 15 min.
-
32.
Put these coverslips in the bottom of 24-well plates for growing HEK293T cells.
-
33.
Add 500 μL of 1% gelatin to each well for coating the coverslip for 30 min at 37°C in CO2 incubator. After 30 min, remove the gelatin and wash the coverslip twice with 1×-PBS (can be stored at 25°C for a week).
-
34.
Seed approximately 20,000 HEK293T cells/coverslip/well for 16 h and keep the plate in CO2 incubator.
-
35.
Next day, transfect the HEK293T cells (∼40% confluent) with the reporter construct 500 ng/mL (either the anti-CSP IB cloned in pLenti-GFP or pLenti-GFP alone).
-
36.
After 24 h of transfection, change the medium, re-transfect the cells with the 100 ng/mL SARS-CoV-2 S construct (HIS-tagged), and wait for another 24 h.
Note: Additional negative controls such as only HEK293T cells seeded onto coverslips or HEK293T cells transfected with anti-CSP IB construct are used.
-
37.
After 24 h of transfection of cells with the SARS-CoV-2 S construct, remove the medium and fix the cells with 4% paraformaldehyde at 25°C for 15 min.
-
38.
Remove the fixation buffer and wash the coverslips three times with 1×-PBS. Then, gently add 500 μL of ice-cold 100% methanol for 10 min and put the coverslips at -20°C to permeabilize the fixed cells. Wash once with 1×-PBS.
-
39.
Add 500 μL of freshly prepared blocking solution (5% BSA and 0.3% Triton X-100 in 1×-PBS) for 1 h at 25°C.
-
40.
Add antibody mix in the blocking solution containing mouse anti- 6×(HIS) antibody (Invitrogen Rockford USA: 4A12E6) at 1:100 dilutions for staining the 6×HIS-tagged SARS-CoV-2 S protein, rabbit anti-calnexin antibody (CST, Beverly, Massachusetts, USA: 52533S) at 1:100 dilution for staining ER and rabbit anti-golgin antibody (Sigma, USA: PA5-52841) at 1:100 dilution for staining Golgi complex. Staining for ER and Golgi complex is performed in separate samples. Keep the cells at 4°C for 16 h in the dark.
-
41.
Next day, wash the cells three times with 1×-PBS and add a secondary antibody mix containing goat anti-mouse IgG Alexa fluor 647 in 1:100 dilution (ThermoFisher Scientific, Waltham, Massachusetts: A21245) for detecting 6×HIS-tagged protein, goat anti-rabbit IgG Alexa Fluor 568 in 1:100 dilution (ThermoFisher Scientific, Waltham, Massachusetts: A11036) for detecting calnexin and golgin protein.
-
42.
Add Hoechst stain (ThermoFisher Scientific, Waltham, Massachusetts) along with the secondary antibodies and incubate cells for 1 h at 25°C and then wash three times with 1×-PBS.
-
43.
Fix the cells on slides using fluoromount-G (Sigma, USA) and keep in dark at 25°C for drying for at least 12 h.
CRITICAL: Rinse the slides with Milli-Q water twice and then with 70% ethanol four times before fixing.
-
44.
Acquire images using a confocal microscope (in our case, an SP8 upright confocal microscope, Leica). All the confocal image stacks were obtained and analyzed using ImageJ software (Results are explained in Figure 3).
Pause point: Stained slides can be stored in dark at 4°C for further screening using confocal microscopy.
Figure 3.
Analyzing the localization of SARS-CoV-2 S protein in control and anti-CSP IB-expressing HEK293T cells during the generation of LV(CoV-2 S) pseudoviruses
HEK293T cells were seeded onto coverslips and incubated for 16 h. The cells were transfected with the anti-CSP sdAb constructs for intracellular expression or with the control pLenti-GFP construct. Thereafter, the cells were re-transfected with the SARS-CoV-2 S construct. After 16 h of re-transfection, the cells were fixed, permeabilized, and stained with ER marker (calnexin) and SARS-CoV-2 S protein carrying a 6×(HIS)-tag using anti-6×(HIS) antibody and anti-mouse Alexa flour 647 as the secondary antibody. The presence of SARS-CoV-2 S protein in ER was measured using SP8 upright confocal microscope (Leica). All the images were analyzed using ImageJ software. The middle panel shows staining of SARS-CoV-2 S along with the transfected cells as indicated for the respective image while in the lower panel the cells were previously transfected to express anti-CSP IB. Nuclear staining is performed by Hoechst stain, pLenti-GFP (control) and anti-CSP IB (sdAb in- fusion with GFP) staining marked by GFP, anti-calnexin antibody were used as ER marker. The upper panel shows all the controls. Scale bar: 160μm.
Assessing the LV(CoV-2 S) pseudoviral particles produced by the anti-CSP IB-expressing HEK293T cells and their control counterparts by immunoblotting
Timing: 3–5 days (for steps45–56)
Here we describe a method to detect differences in the levels of spike protein produced by the transfected HEK293T cells in the presence or absence of intrabody though western blot.
-
45.
Take the transfected HEK293T cells from the described conditions and add 1 mL C-100 buffer (can be stored at 25°C). Vortex for 3 min and keep at −80°C for 16 h.
Note: For making cell lysates, we used 10, 00,000 cells/mL of C-100 buffer.
-
46.
Next day, take out the tubes from −80°C and thaw the samples at 4°C followed by vortexing for five times (30 s vortex and then 1 min on ice) in 7 min.
-
47.
Then centrifuge the tubes at 16,000 × g for 20 min at 4°C.
-
48.
Collect the supernatant and estimate the concentration of protein. Normalize the concentration in different samples.
-
49.
Resolve the prepared samples using a 12% SDS-PAGE and transfer the resolved polypeptides onto PVDF membrane using a wet transfer method for 1.5 h at 90V at 4°C.
-
50.
Soak the PVDF membrane in 5% blocking solution (prepare fresh every time) (5% skim milk in 10 mL of 0.05% PBST) for 16 h at 4°C.
-
51.
Next day, wash the membrane three times with 0.05% PBST solution, each for 5 min at a rocker.
-
52.
After washings, add mouse anti-6×HIS monoclonal antibody (1:5000 dilutions in 0.05% PBST solution) for 2 h at 25°C on a rocker.
-
53.
Wash the blots five times with 0.1% PBST, incubating the membranes for 10 min on a rocker.
-
54.
Add goat anti-mouse IgG conjugated with alkaline phosphatase (ALP) in 1:10000 dilutions with 0.1% PBST for 1 h at 25°C at a rocker.
-
55.
Wash the membrane five times on a rocker with 0.1% PBST solution, each for 10 min.
-
56.
Immerse the membrane in femtolucent (stored at 4°C) for 2–3 min and then expose it to X-Ray film in the dark room (Schematic is shown in Figure 4 and results are shown in the original article Figure 5B).
Figure 4.
Schematic for the production of LV(CoV-2 S) pseudoviruses from transfected HEK293T cells (control and anti-CSPIB condition) and measuring the spike production in the cell lysate of HEK293T cells using western blotting
Surface and intracellular expression of SARS-CoV-2 S protein in anti-CSP IB-expressing or the control HEK293T cells
Timing: 3–4 days (for steps57–64)
In this section, we measure the expression of SARS-CoV-2 S protein in transfected HEK293T cells in the absence or presence of the intrabody through flow cytometry. We describe the use of flow cytometry in detecting SARS-CoV-2 S protein using anti-CSP sdAb.
-
57.
Transfect control or the anti-CSP IB-expressing HEK293T cells to produce LV(CoV-2 S) based pseudoviruses.
-
58.
After 72 h of transfection, scrap out the HEK293T cells and wash twice with 1×-PBS.
-
59.
To approximately one million HEK293T cells, add 10 μg/mL of anti-CSP sdAb and incubate the cells for 30 min at 25°C to assess the surface expression of the SARS-CoV-2 S protein.
-
60.
To detect intracellular SARS-CoV-2 S protein, fix the cells with 100 μL of fixation buffer (IC Fixation buffer, Invitrogen, stored at 4°C) for 30 min on ice.
-
61.
Wash cells with 1×-permeabilization buffer {(diluted in Milli-Q water (1:1), dilute before every experiment} (Permeabilization buffer 10×, Invitrogen, stored at 4°C) and then incubate the cells with the permeabilization buffer for 20 min on ice.
-
62.
After incubation, add anti-CSP sdAb (10 μg/mL) for another 30 min on ice. Wash cells twice with 1× permeabilization buffer.
-
63.
Add (1:200 dilution) of streptavidin-APC for 30 min at 25°C. Wash the cells twice with 1× permeabilization buffer.
-
64.
After washing resuspend the cells in 200 μL of permeabilization buffer, acquire using a flow cytometer (Results of differential expression of the SARS-CoV-2 S protein in the above two conditions are explained in Figure 5).
Expected outcomes
This protocol can be used to express sdAbs intracellularly, such as the one recognizing polybasic cleavage site contained in SARS-CoV-2 S protein (anti-CSP IB) in HEK293T cells. Since the anti-CSP IB is produced as a fusion product with GFP and c-Myc tag, assessing its expression, colocalization with the SARS-CoV-2 S protein, and eventually determining the fate of the viral protein can be analyzed using different approaches. Furthermore, the protocol described here can be used to reveal events in the viral assembly and biogenesis and the role of different subcellular compartments in the process. A stable cell line of anti-CSP IB-expressing HEK293T and Vero E6 cells can be made similarly to assess the viral biogenesis processes of different viruses with similar recognition sequences: RRxR. Some of the examples of such viruses include SARS-CoV-2, HSV1, PPRV, and dengue viruses. Furthermore, the scope of the described protocol could be extended to analyze the functionality of intrabodies targeting different viral proteins and assessing events in the biogenesis of other viruses or intracellular pathogens.
Limitations
We have not tested the viral assembly, biogenesis, and infectivity of SARS-CoV-2 S or any other live virus in the cells expressing the anti-CSP IB at present. The protocol was applied to evaluate the biogenesis of pseudovirus expressing SARS-CoV-2 S protein in the control and anti-CSP IB-expressing cells.
Troubleshooting
Problem 1
No detectable spike protein in HEK293T cells supernatant 72 h post-transfection.
Potential solution
Please check the quality and concentration of all the plasmids used for pseudovirus production using nanodrop (A260/A280 readings before each transfection reaction). Also monitor the transfected cells phenotypically.
Problem 2
No detectable amount of LV(CoV-2 S) based pseudoviruses from control transfected HEK293T cells.
Potential solution
The transfected cells could have crossed Hayflick limit, wherein the somatic cells stop dividing. So, revive new vial of HEK293T cells for further transfection.
Problem 3
Detachment of HEK293T cells post transfection.
Potential solution
Removal of extra PEI from the transfected HEK293T cells is of critical after 6 h of transfection to avoid the detachment and death of transfected HEK293T cells.
Resource availability
Lead contact
All the queries regarding reagents and resource availability will be fulfilled by the lead contact, Dr. Sharvan Sehrawat (sharvan@iisermohali.ac.in).
Materials availability
The anti-CSP sdAb expression and intrabody constructs generated as part of this study will be made available upon request to lead contact.
Acknowledgments
The study was supported partly by intramural funding from IISER Mohali, extramural grants from Department of Biotechnology (DBT) numbered BT/PR20283/BBE/117/2016, and from Department of Science Technology (DST) numbered IPA/2021/000136 to S. Sehrawat. S. Singh received a fellowship from the Indian Council of Medical Research, and S.D. received a fellowship from the intramural fund of IISER Mohali. All the images were prepared using BioRender software subscribed by Ayush Jain. The author would also like to thank Azeez Tehseen for reviewing the manuscript.
Author contributions
Conceptualization: S. Sehrawat, S.D., S. Singh; methodology: S.D., S. Singh; writing: S. Sehrawat, S.D., S. Singh; supervision: S. Sehrawat; funding acquisition: S. Sehrawat.
Declaration of interests
The authors declare no competing interests.
Data and code availability
No data or code were developed during this study.
References
- 1.Singh S., Dahiya S., Singh Y.J., Beeton K., Jain A., Sarkar R., Dubey A., Tehseen A., Sehrawat S. Robust anti-SARS-CoV2 single domain antibodies cross neutralize multiple viruses. iScience. 2022;25:104549. doi: 10.1016/j.isci.2022.104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paganetti P., Calanca V., Galli C., Stefani M., Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer’s Abeta peptide. J. Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur M., Dubey A., Khatri M., Sehrawat S. Secretory PLA2 specific single domain antibody neutralizes Russell viper venom induced cellular and organismal toxicity. Toxicon. 2019;172:15–18. doi: 10.1016/j.toxicon.2019.10.240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data or code were developed during this study.