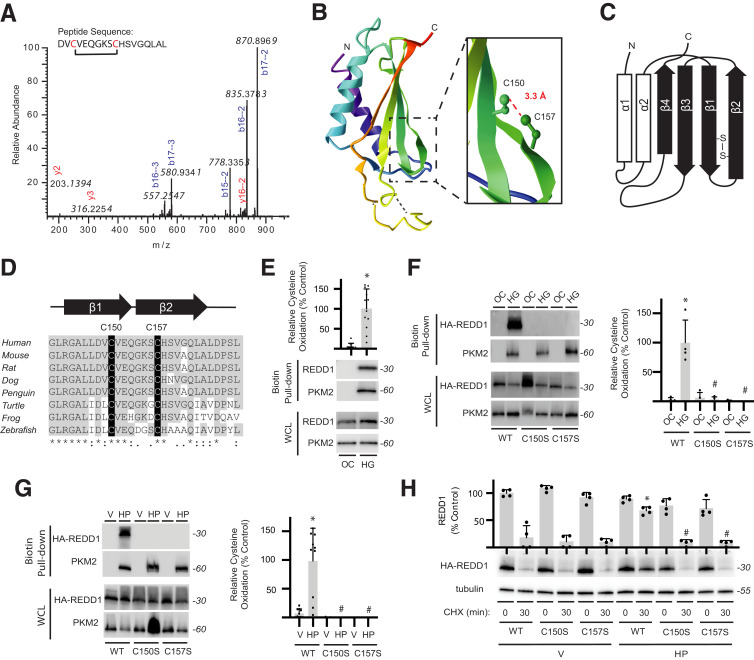
Figure 3.
REDD1 degradation is reduced by a redox-sensitive disulfide bond. A: Liquid chromatography–tandem mass spectrometry fragmentation of AspN-digested FLAG-REDD1 identified a cystine-linked peptide by loss of 2.01565 Da that included C150/C157. B: REDD1 protein structure (PDB ID 3LQ9) includes an α/β sandwich with two antiparallel α-helices and four β-strands. Inset highlights 3.3-Å distance between the C150 and C157 side chains. C: Two-dimensional graphic of REDD1 secondary structure illustrates location of C150/C157 cross-strand disulfide. D: C150/C157 of REDD1 exhibit sequence conservation. E: REDD1 cysteine oxidation in MIO-M1 cells exposed to medium supplemented with 30 mmol/L glucose (HG) or an osmotic control (OC) for 4 h was evaluated by biotin switch assay. Western blotting was used to evaluate REDD1 and PKM2 in biotin pull-downs and whole-cell lysates (WCL). Representative blots are shown. Protein molecular mass in kDa is indicated at right of blots. REDD1 in biotin pull-down was quantified. PKM2 was evaluated as a positive control. F and G: WT, C150S, and C157S HA-REDD1 variants were expressed in REDD1-KO MIO-M1 cells. Cells were exposed to OC vs. HG for 4 h (F) or to vehicle (V) or H2O2 (HP) for 2 h (G). REDD1 cysteine oxidation was evaluated by biotin switch assay. H: WT, C150S, and C157S HA-REDD1 variants were expressed in REDD1-KO MIO-M1 cells. HA-REDD1 variant degradation was evaluated by cycloheximide (CHX)-chase assay. Data are represented as mean ± SD. *P < 0.05 vs. OC or Veh; #P < 0.05 vs. WT.