FIGURE 2.
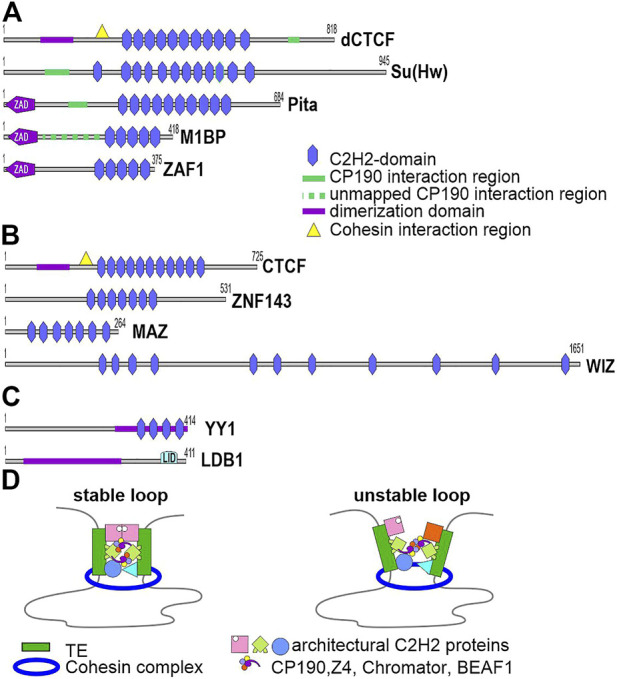
Main Drosophila C2H2 architectural proteins and mammalian proteins involved in long-range interactions. (A) Schematic structures of the Drosophila C2H2 architectural proteins studied in most detail: dСTCF, Su(Hw), Pita, M1BP, and ZAF1. Unstructured regions interacting with CP190 were precisely mapped in CTCF, Su(Hw) and Pita, but not in M1BP. (B) Schematic structures of human CTCF and the C2H2 proteins ZNF143, MAZ, and WIZ, which were shown to cooperate with CTCF in organizing the chromosome architecture. A motif responsible for interactions with the cohesin complex was mapped only in CTCF. (C) LDB1 and YY1, which maintain specific enhancer–promoter interactions in mammals. Homodimerization domains are indicated. (D) Currently, there are no experimental data showing that cohesin-mediated loop extrusion and protein-protein interactions between transcription factors cooperatively form chromatin loops between regulatory elements. However, a model can be proposed according to which architectural C2H2 proteins with associated transcription factors form regulatory elements that limit cohesin-mediated loop extrusion through multiple interactions with different cohesive subunits. Homodimerization between C2H2 architectural proteins and interaction between associated transcription factors stabilize the chromatin loop.
