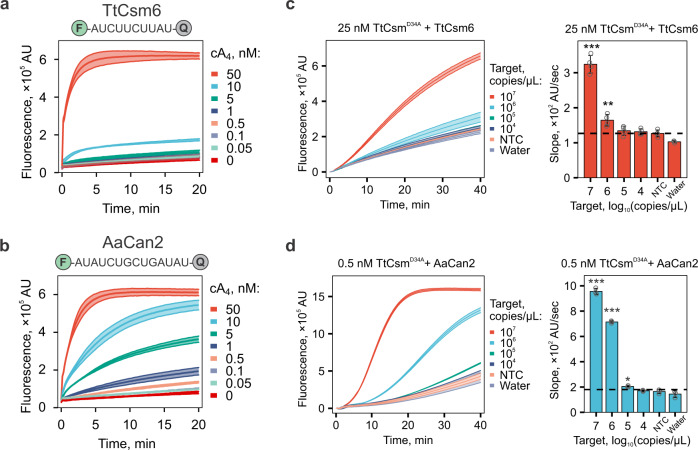
Fig. 3. Incorporation of Can2 nuclease improves sensitivity of Csm-based RNA detection.
a TtCsm6 (300 nM) and b AaCan2 (300 nM) cleavage assays with fluorescent ssRNA reporter (top) in the presence of varying cA4 activator concentrations (shown with colors). Data are shown as the mean (center line) of three replicates ± S.D. (ribbon). The optimal fluorescent reporter (top) was determined using RNA library screen in Supplementary Fig. 6. TtCsm RNA detection coupled with TtCsm6- (c) and AaCan2-based (d) assays were performed using samples with target RNA concentrations ranging from 107 to 104 copies/µL. Samples were prepared by spiking IVT fragments of SARS-CoV-2 N gene into total RNA extracted from nasopharyngeal swab patient sample negative for SARS-CoV-2. Cleavage of fluorescent RNA reporter was detected by measuring fluorescence every 10 s in a real-time PCR instrument (left). Data were plotted as mean of four replicates. Simple linear regression was used to calculate slopes for linear regions of the curves. Bars show mean values (n = 4) ± S.D. (right). Data was analyzed with one-way ANOVA followed by multiple comparisons to NTC sample using one-tailed post-hoc Dunnett’s test. ***p < 0.001; **p < 0.01; *p < 0.05. AU = arbitrary units. Source data are provided as a Source Data file.