Figure 5.
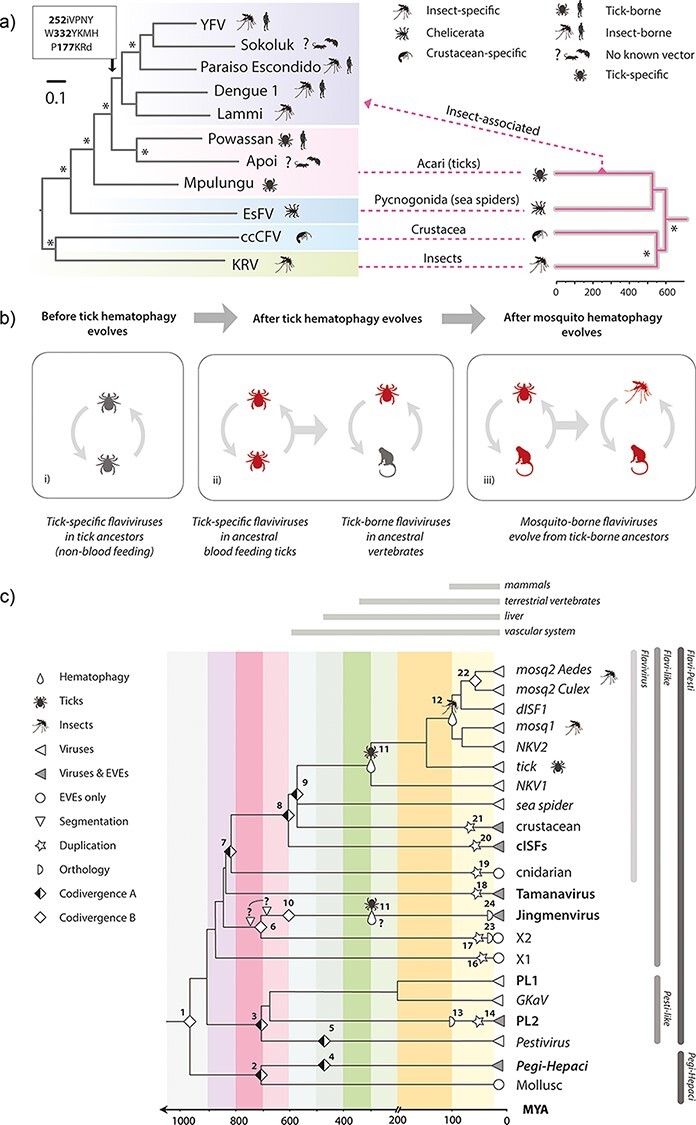
The timeline of flavivirus evolution. (a) Evolution of arthropod-borne flaviviruses. The phylogeny shown on the left was constructed from an alignment spanning 804 residues of the precursor polyprotein (substitution model = LG likelihood). The phylogeny on the right is a time-calibrated phylogeny of arthropod hosts/vectors of classical flaviviruses, obtained via TimeTree (Kumar et al. 2017). The figure shows the hypothesised relationships between the host and virus trees, with virus phylogeny broadly following host phylogeny at higher taxonomic ranks but diverging dramatically from this pattern when tick-borne viruses of vertebrates emerge. (b) Proposed model for the origin and evolution of the vector-borne flaviviruses. Three stages are shown: (i) an ancestral group of non-vectored tick viruses is present; (ii) tick haematophagy provides prolonged exposure to vertebrate blood, thereby affording tick-specific flaviviruses the opportunity, through chance events and mutation, to acquire a capacity to replicate in vertebrate cells and ultimately become tick-borne flaviviruses of vertebrates; (iii) the existence of tick-borne flaviviruses occasions viraemic vertebrate hosts, exposing mosquitoes and other haematophagous insects to flaviviruses via blood-feeding and ultimately allowing them to acquire a vector role.(c) A model for long-term evolution of flavivirids. A time-scaled tree summarising the phylogenetic relationships of flavivirids and calibrated using information obtained from analysis of EVEs, co-phyletic analysis, and the fossil record of haematophagous arthropods. Symbols on the phylogenies indicate types of calibration as shown in the key. Numbers adjacent to symbols link to rows in Table 3. ‘Codivergence type A’ = codivergence supported by co-phylogeny. Codivergence type B = potential codivergence-based calibrations without supporting evidence. Vertical lines to right of the tree show taxonomic groups.
