Figure 1. Interaction network of the IFT‐B complex obtained by chemical cross‐linking and mass spectrometry.
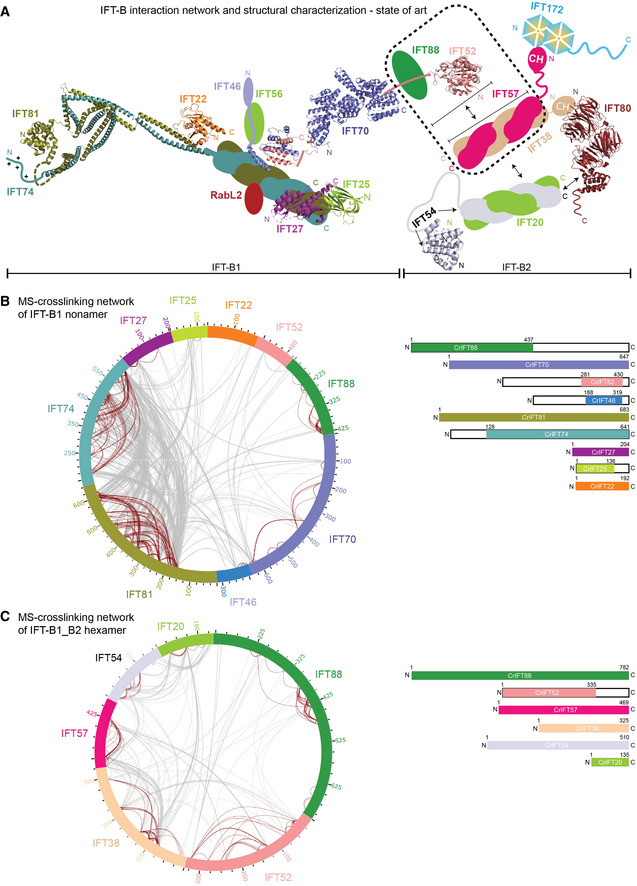
- Schematic representation of IFT‐B complex architecture based on published structural and biochemical data. Structural information is available for Trypanosoma brucei IFT22 and the N‐termini of IFT81/74 (PDB: 6ian), the Chlamydomonas reinhardtii IFT27/25 heterodimer (PDB: 2yc2), the C‐termini of IFT52 and IFT46 from Tetrahymena thermophila (PDB: 4uzz), the IFT70/52 complex from Chlamydomonas reinhardtii (4uzy), the globular N‐terminal GIFT domain of IFT52 from Chlamydomonas reinhardtii (PDB: 5FMR) as well as from Mus musculus (PDB: 5FMS), the N‐terminal CH‐domain of IFT54 from Mus musculus (5FMU) and IFT80 from Chamydomonas reinhardtii (PDB: 5N4A). The IFT‐B complex is subdivided into biochemically salt stable IFT‐B1 (IFT88/81/74/70/56/52/46/27/25/22/RabL2) and IFT‐B2 (IFT172/80/57/54/38/20) complexes.
- The inter‐ and intramolecular cross‐linking network within the Chlamydomonas IFT‐B1 nonamer (IFT881–437/70/52281–430/46188–319/81/74128‐C/27/251–136/22) are depicted as a cartwheel diagram (left panel). The gray lines show intermolecular cross‐linking pairs, and the brown lines show the intramolecular cross‐linking pairs. The protein constructs of the IFT‐B1 nonamer are indicated on the right.
- The inter‐ and intramolecular cross‐linking network within a CrIFT‐B1‐B2 hexamer (IFT88/521–335/57/38/54/20) is displayed as a cartwheel. In this protein complex, only the C‐terminal part of IFT52 is truncated while all other proteins are full length (see schematics on the right panel).
