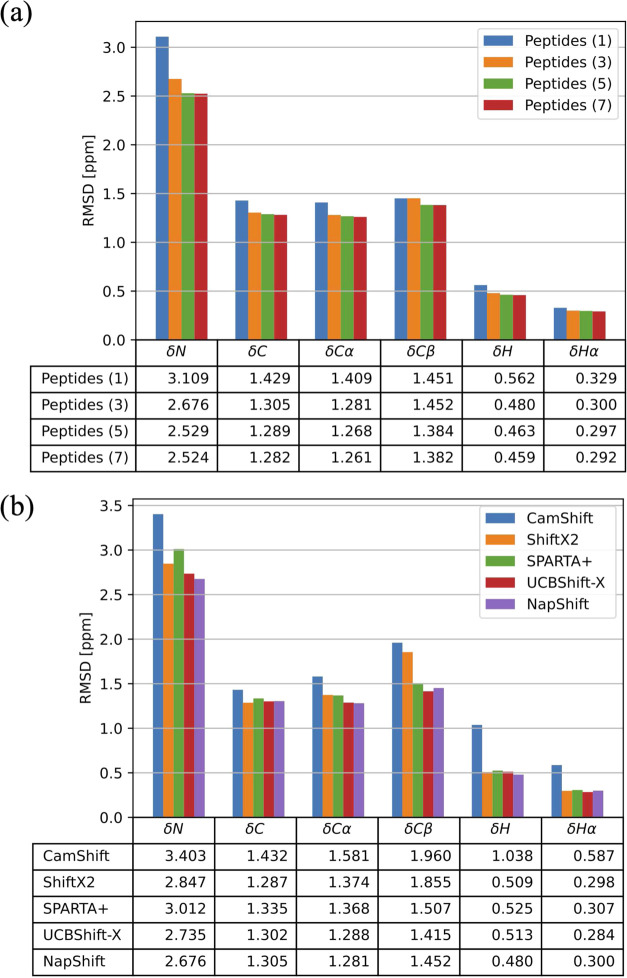
Figure 2.
(a) Comparison of the RMSD in ppm between the NapShift-predicted CS and the experimental CS for varying input peptide lengths (1, 3, 5, and 7) to the NapShift ANN. (b) Comparison of the RMSD between predicted and experimental CS for five different prediction methods. NapShift outperforms most of the prediction methods, for all backbone atoms, on the same set of 250 test PDB structures. In the case of CamShift, a 30 ppm threshold on the CS predictions was applied to avoid systematic errors that would distort the RMSD values.