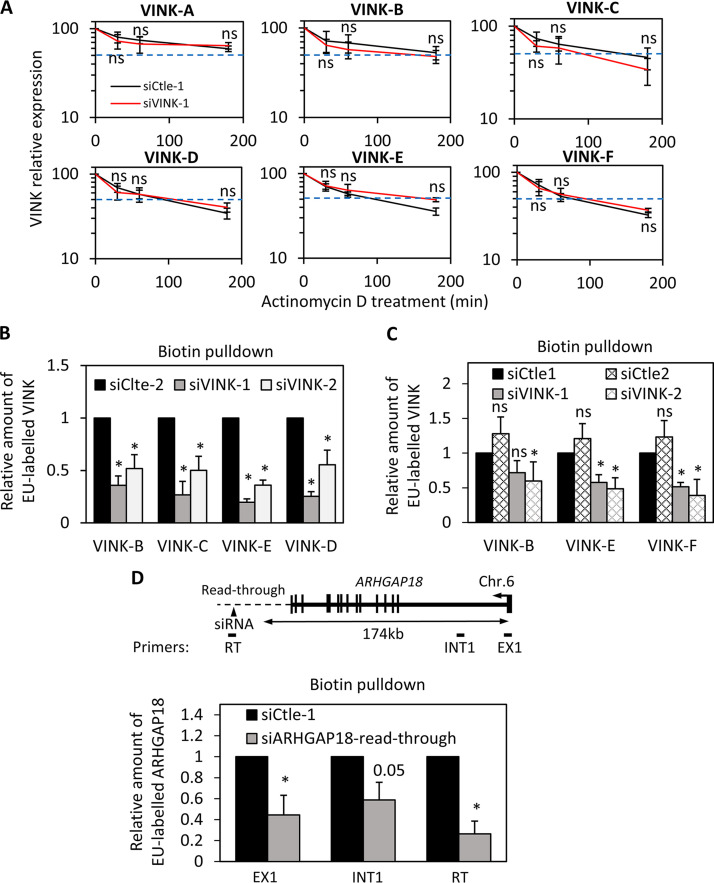
FIG 2.
Small interfering RNAs affect VINK transcription. (A) Senescent WI38-RAF1 cells were transfected with the VINK-1 or control (Ctle-1) siRNAs. At 48 h later, actinomycin D (10 μg/mL) was added or not. Cells were harvested after the indicated times. Total RNA was prepared with TRIzol and analyzed by RT-qPCR for the expression of GAPDH mRNA or VINK (using the indicated primers). The amount of VINK RNA was standardized to GAPDH mRNA (which did not vary during this time course of actinomycin D treatment) and calculated relative to a value of 100 for the untreated sample. The means and standard deviations from 3 independent experiments are shown. Student's t test was performed to compare VINK1 and Ctle-1 siRNA samples at each time point of actinomycin D treatment. ns (nonsignificant), P > 0.1. (B) Senescent WI38-RAF1 cells were transfected with the indicated siRNAs. Nascent RNAs (biotin pulldown) were prepared and analyzed by RT-qPCR for the expression of GAPDH mRNA or VINK (using the indicated primers). The amount of VINK RNA was standardized to GAPDH mRNA and calculated relative to a value of 1 for the Ctle-1 siRNA. The means and standard deviations from 3 independent experiments are shown. *, P < 0.05 compared to Ctle-2 siRNA. (C) Same experiment as in panel B, except that U2OS cells were used. *, P < 0.05 compared to Ctle-1 siRNA; ns, P > 0.1. (D) Senescent WI38-RAF1 cells were transfected with Ctle-1 siRNA or an siRNA directed against ARHGAP18 START RNA (14). Nascent RNAs (biotin pulldown) were prepared and analyzed by RT-qPCR for the expression of GAPDH mRNA or the indicated primers on ARHGAP18. The amount of ARHGAP18 or START RNA was standardized to GAPDH mRNA and calculated relative to a value of 1 for the Ctle-1 siRNA. The means and standard deviations from 3 independent experiments are shown. *, P < 0.05 compared to Ctle-1 siRNA; ns, P > 0.1; P value is shown for the comparison to Ctle-1 siRNA when 0.05 < P < 0.1.