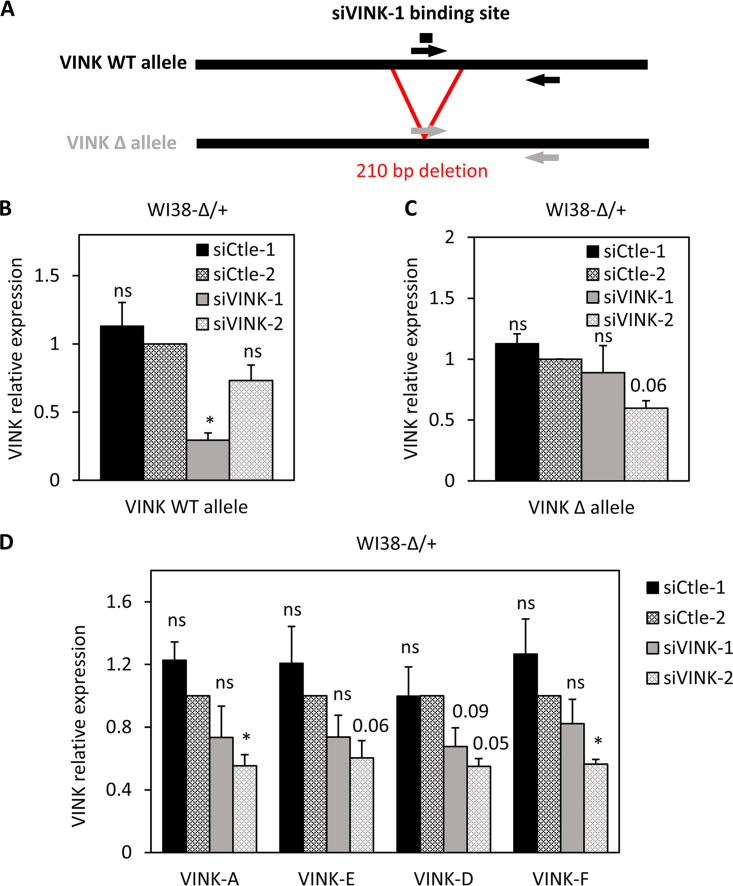
FIG 5.
VINK siRNA requires cis-targeting to chromatin to repress VINK transcription. (A) Schematic representation of the two alleles of VINK present in WI38-Δ/+ with primers allowing their specific detection. The deleted allele was recognized by a primer located on the deletion junction. (B) Senescent WI38-Δ/+cells were transfected with the indicated siRNAs. At 72 h later, total RNA was prepared and analyzed by RT-qPCR for the expression of GAPDH mRNA or the VINK wild-type allele. The amount of VINK allele was standardized to GAPDH mRNA and calculated relative to a value of 1 for cells transfected with the Ctle-2 siRNA. The means and standard deviations from 3 independent experiments are shown. *, P < 0.05 compared to Ctle-2 siRNA; ns, P > 0.1. (C) Same experiment as in panel B, except that the deleted VINK allele was analyzed. The means and standard deviations from 3 independent experiments are shown. *, P < 0.05 compared to Ctle-2 siRNA; ns, P > 0.1; the P value is shown for the comparison to Ctle2 siRNA when 0.05 < P < 0.1. (D) Same experiment as in panels B and C, except that total VINK expression was analyzed with the indicated primers. The means and standard deviations from 3 independent experiments are shown. *, P < 0.05 compared to Ctle-2 siRNA; ns, P > 0.1; the P value is shown for the comparison to Ctle-2 siRNA when 0.05 < P < 0.1.