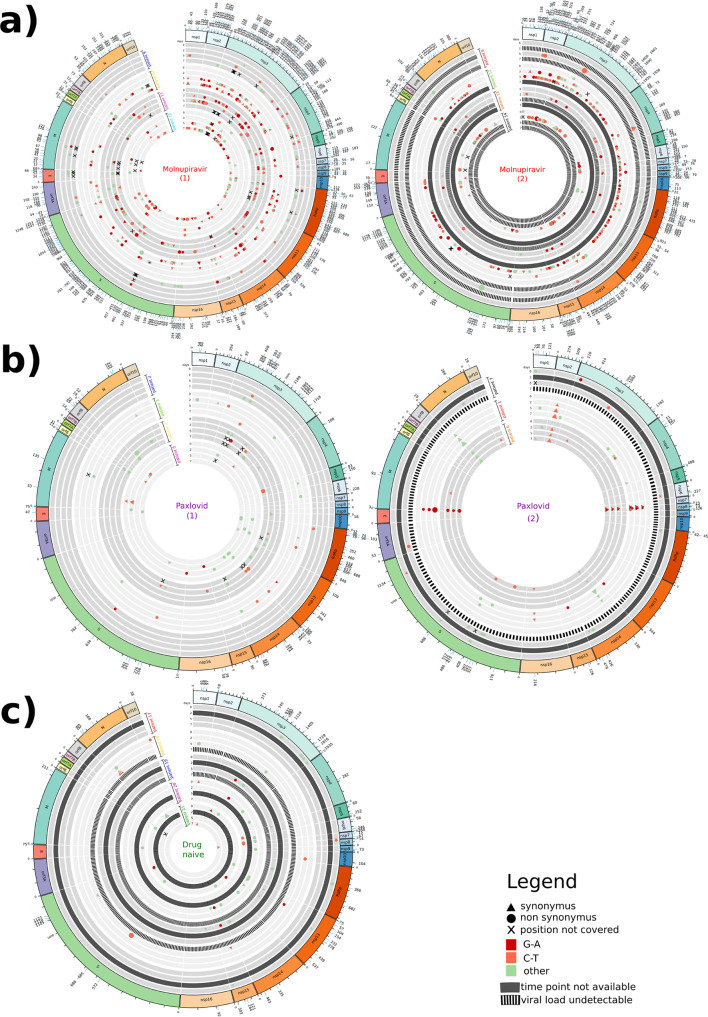
Fig. 3. Circos plots representing the SNPs found in SARS-CoV-2 strains.
Panels represent SARS-CoV-2 strains found in (a) Molnupiravir-treated, (b) Paxlovid-treated and (c) drug-naïve individuals across time points. Each time point is defined by one track and grouped according to patient. Tracks in black represent timepoints not available; striped tracks represent samples not sequenced as viral load was undetectable. SNPs are defined by different colors, shapes and size according to type of nucleotide substitution (red: G-A; orange: C-T; green: other), non-synonymus (circle) or synonymus mutations (triangle), and intra-patient prevalence, respectively. The outside track corresponds to SARS-CoV-2 genome architecture. Overlapping orfs are not reported and comprise: orf2b (coordinates 21744–21860) overlapping S; orf3b (coordinates 25814–25879), orf3c (coordinates 25457–25579), orf3d (coordinates 25524–25694) and orf3d-2 (coordinates 25596–25694) overlapping orf3a; orf9b (coordinates 28284–28574) and orf9c (coordinates 28734–28952) overlapping N.