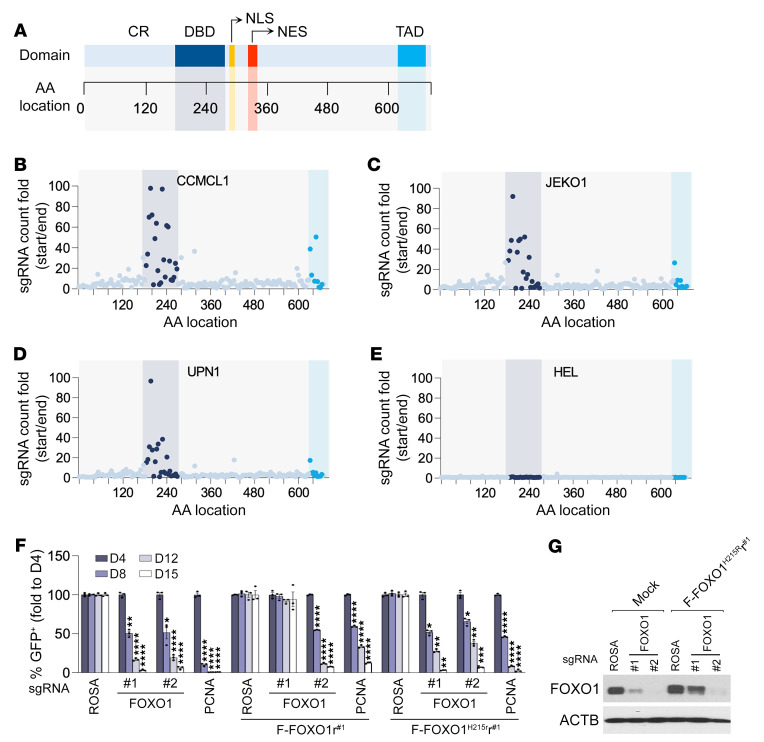
Figure 4. DBD and TAD of FOXO1 are required for its MCL lineage-survival function.
(A) A schematic graph of structural domains of FOXO1 protein. CR, conserved region; DBD, DNA binding domain; TAD, transactivation domain; NLS, nuclear localization sequence; NES, nuclear export sequence. (B–E) Systematic evaluation of 167 FOXO1 sgRNAs in negative selection experiments. The location of each sgRNA relative to the FOXO1 protein is indicated along the x axis. The y axis is the fold change of the abundance of individual sgRNAs (ratio of start to end point) in Cas9-expressing CCMCL1 (B), JEKO1 (C), UPN1 (D), and HEL (E) cells after 14 population doublings. (F) Competition-based proliferation assays of sgRNAs against FOXO1 in Cas9-transduced CCMCL1 cells expressing wild-type FOXO1 (FOXO1r#1) or DNA binding–defective mutant (FOXO1H215Rr#1). The FOXO1r#1 and FOXO1H215Rr#1 mutants are CRISPR-resistant to the action of sgFOXO1#1 but sensitive to sgFOXO1#2. Data represent mean ± SEM (n = 3). Results are representative of 3 independent experiments. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple-comparison test. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001. (G) Immunoblot analysis of control or FOXO1H215Rr#1-expressing CCMCL1 cells at day 3 after transduction of indicated sgRNAs.