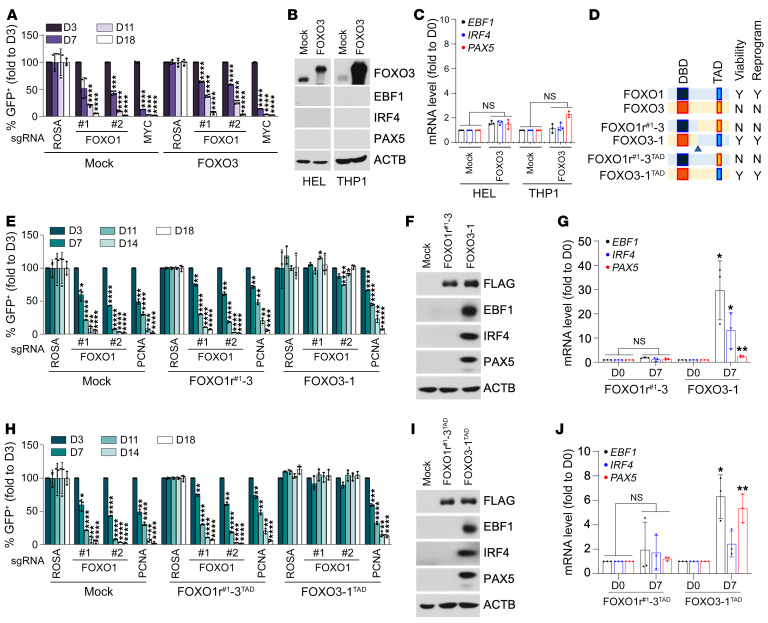
Figure 5. TAD of FOXO1 specifies its MCL lineage–supporting activity.
(A) Competition-based proliferation assays for indicated FOXO1-targeted sgRNAs in mock control or FOXO3-transduced CCMCL1 cells. Data represent mean ± SEM (n = 4). (B and C) Immunoblot (B) and RT-qPCR analysis (C) of EBF1, IRF4, or PAX5 expression in FOXO3-transduced HEL or THP1 cells. Cell lysates and total RNA were prepared at day 20 after infection of FOXO3-encoding lentivirus. Data represent mean ± SEM (n = 3). (D) A schematic of domain-swapped FOXO1 and FOXO3 mutants. N, no; Y, yes. (E) Competition-based proliferation assays for indicated FOXO1-targeted sgRNAs in FOXO1r-3–transduced (left) or FOXO3-1–transduced (right) CCMCL1 cells. Data represent mean ± SEM (n = 3). (F and G) Immunoblot (F) and RT-qPCR (G) analysis of EBF1, IRF4, and PAX5 induction in FOXO1r#1-3–transduced (left) or FOXO3-1–transduced (right) THP1 cells. Cell lysates and total RNA were prepared at day 7 after infection of lentivirus encoding indicated variants. Data represent mean ± SEM (n = 3). (H) Competition-based proliferation assays in FOXO1r#1-3TAD–transduced (left) or FOXO3-1TAD–transduced (right) CCMCL1 cells. Data represent mean ± SEM (n = 3). (I and J) Immunoblot (I) and RT-qPCR (J) analysis of EBF1, IRF4, and PAX5 induction in FOXO1r#1-3TAD–transduced (left) or FOXO3-1TAD–transduced (right) THP1 cells. The FOXO1r#1-3TAD variant is CRISPR-resistant to the action of sgFOXO1#1 but remains sensitive to sgFOXO1#2. Data represent mean ± SEM (n = 3). (A, C, E, G, H, and J) Results are representative of 3 independent experiments. Statistical analysis was performed using 1-way ANOVA with Tukey’s multiple-comparison test in A, E, and H and using 2-tailed unpaired Student’s t test in C, G, and J. *P < 0.05, **P < 0.001, ***P < 0.0005, ****P < 0.0001.