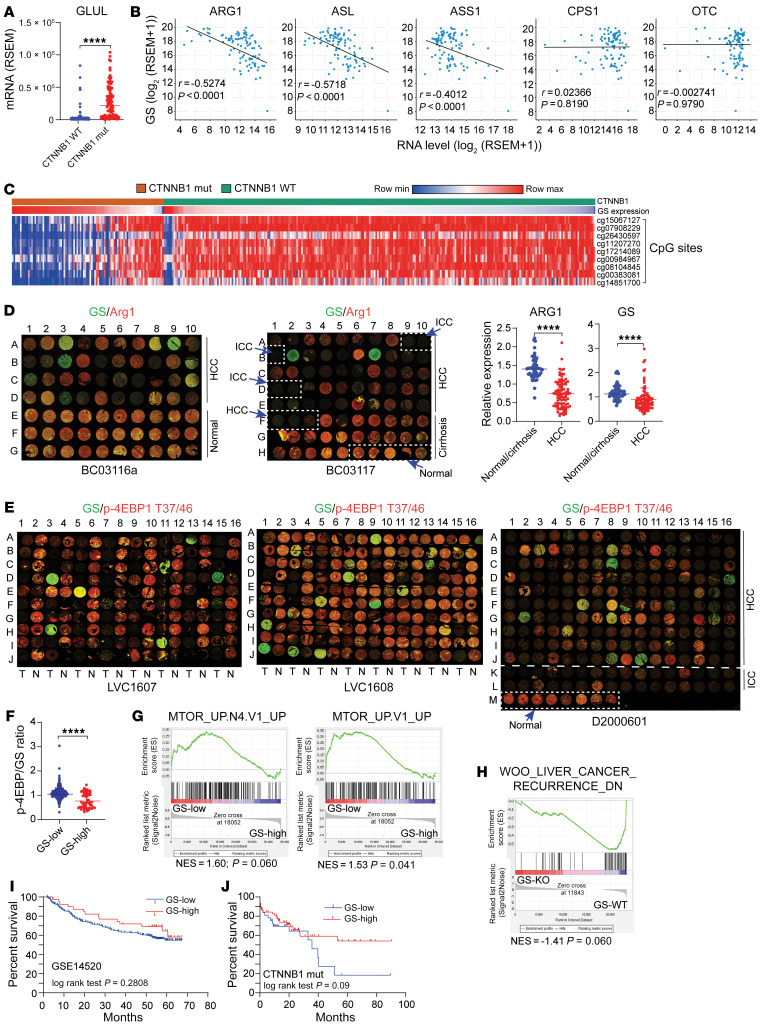
Figure 9. Defective nitrogen waste removal in HCC clinical samples.
(A) Relative GLUL mRNA levels in liver cancer patients with WT or mutated CTNNB1 (TCGA) were compared and are expressed as mean ± SEM. (B) Correlations between hepatic GLUL and UCE expression in patients with CTNNB1 mutations (TCGA) was calculated by Pearson’s correlation. (C) CTNNB1 mutation correlates positively with hepatic GS expression but inversely with GLUL CpG island methylation. (D) Two liver TMAs were costained for GS (green) and ARG1 (red) and quantified by ImageJ and normalized to the respective median (right panel). (E) TMAs with HCC tumors (T), adjacent normal tissue (N), or intrahepatic cholangiocarcinoma (ICC) were costained for GS (green) and p-4EBP1 T37/46 (red). (F) Ratios of p-4EBP1 T37/46 to GS intensities were plotted. Results are expressed as mean ± SEM. The cutoff value for GS-high (n = 52) versus -low (n = 372) was determined using the best separation between the 2 modes of a bimodal distribution. ****P < 0.0001 by 2-tailed t test (A, D, and F). (G) GSEA enrichment plots show that mTORC1 target genes correlate inversely with GS level in CTNNB1-mutated patients (TCGA). The cutoff value for GS-high (n = 62) versus -low (n = 34) was determined using the best separation between the 2 modes of a bimodal distribution. P value was calculated by estimated score in GSEA. (H) The TCGA data of the indicated study were stratified into the RNA-seq results obtained from WT and Glul-KO mice 2 weeks after c-Met/ΔN90-β-catenin injection (n = 4 in each group). GSEA plots show that high GS expression correlates with low recurrence. (I) Kaplan-Meier survival curves of patients with high (n = 41) versus low (n = 201) hepatic GS expression (GSE14520; P = 0.28, log rank). (J) In HCC patients harboring CTNNB1 mutations (TCGA) as shown in G, high GS expression tends to associate with better survival (P = 0.09, log rank).