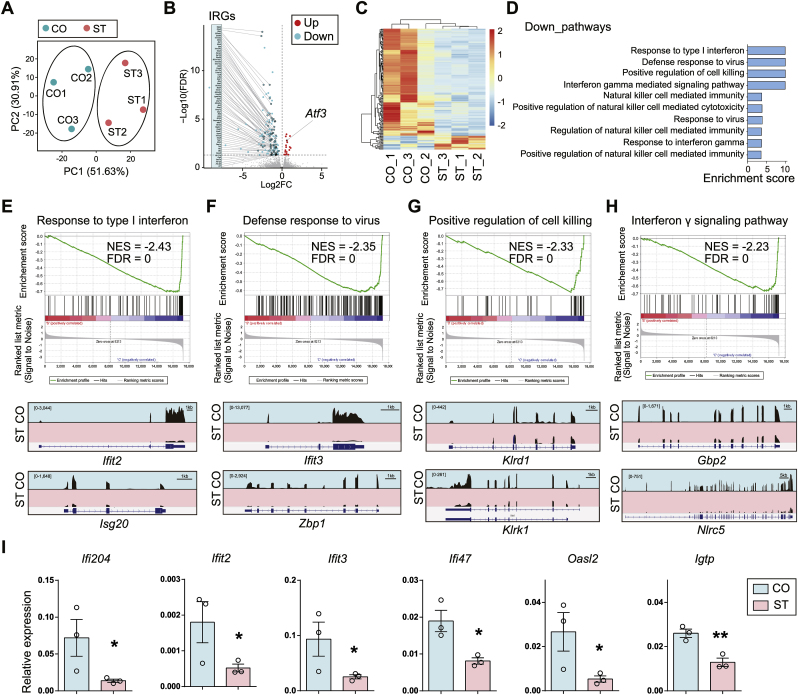
Fig. 2.
Microglia-specific transcriptional repression of genes in IFN-signaling pathway after chronic stress. (A) PCA plot of RNA-seq using isolated microglia from mice exposed to chronic stress (ST) and controls (CO). N = 3/group. (B) The volcano plot shows the differential gene expression in ST compared to CO. Red dot, up-regulated genes (Up); blue dot, down-regulated genes (Down); gray dot, non-significant genes. FDR <0.05. Notice the massive down-regulation of genes in ST, and many of them (65 out of 140) are IFN-regulated genes (IRGs) (Interferome v2.0). Notice the increase of Atf3 in ST. (C) Clustered heatmap shows differential gene expression (ST v.s. CO). (D) Gene Set Enrichment Analysis (GSEA) shows down-regulated functional pathways (ST vs. CO). Notice high enrichment for IFN-signaling pathways. Enrichment score = -Log10(P+10−10). (E–H) (Top) Enrichment plots and (Bottom) IGV map tracks for representative genes in the top four down-regulated pathways. Notice a decrease of RNA-seq signal for IRGs in ST compared to CO. (I) Real-time RT-PCR validation for the down-regulated IRGs. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)