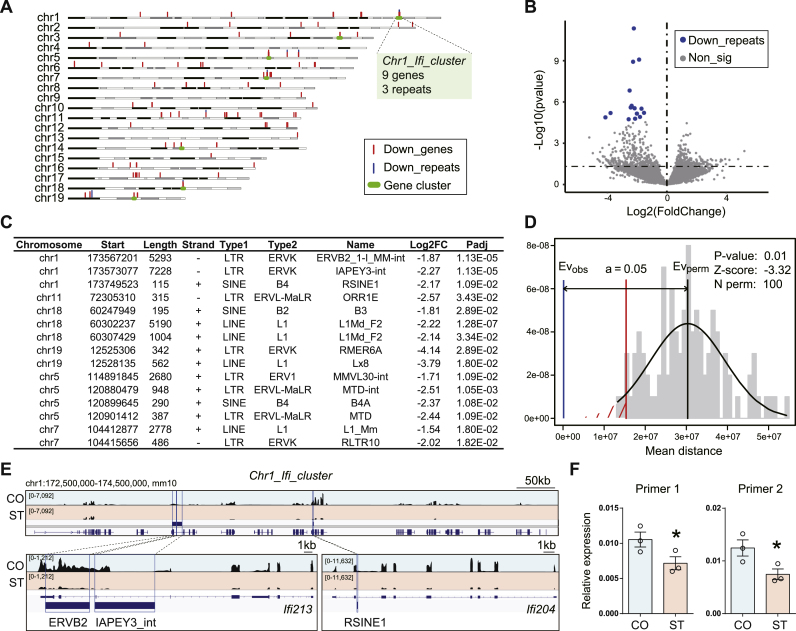
Fig. 3.
Microglia-specific transcriptional repression of DNA repeats after chronic stress. (A) Genome distribution of down-regulated genes (down_genes, red vertical bars) and DNA repeats (blue vertical bars). Gene clusters are highlighted with green horizontal bars. Notice the cluster of genes encoding IFN-induced proteins located at the end of chromosome 1 (“Chr1_Ifi_cluster”). (B) The volcano plot shows differential DNA repeats transcription after chronic stress (ST v.s. CO). Blue, down-regulated DNA repeats (“Down_repeats”); gray, non-significant. FDR <0.05. N = 3/group. The genome locations of these 15 “Down_repeats” are labeled in panel A. (C) List of significantly down-regulated DNA repeats. (D) Permutation test for the distance between significantly down-regulated DNA repeats and genes. (E) IGV map tracks show RNA-seq signal at Chr1_Ifi_cluster. Notice three DNA repeats, ERVB2, IAPEY3_int, and RSINE1, located in the gene bodies of Ifi213 and Ifi204. (F) Real-time RT-PCR validation for the ERVB2 in Chr1_Ifi_cluster using two different primers. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)