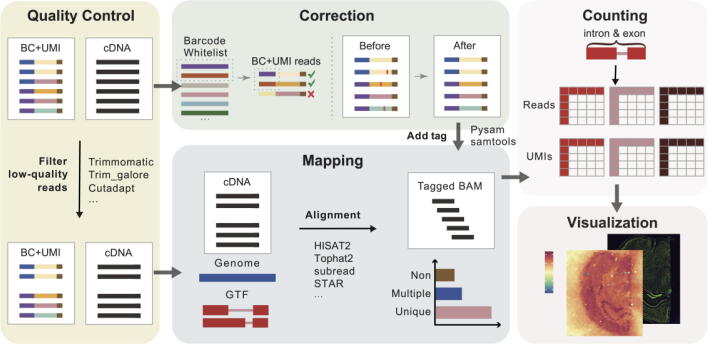
Fig. 1.
Framework of the typical spatial transcriptome preprocessing pipeline. First, FASTQ files are subjected to quality control treatment. Reads with low-quality barcodes or UMIs are discarded, and the remaining reads are mapped to the corresponding genome. Next, a barcode/UMI correction process is carried out to fetch reads of the same origin. Then, a spot-by-gene count matrix is generated according to cDNA and barcode/UMI reads. Finally, the expression matrix is visualized as a series of heat maps.