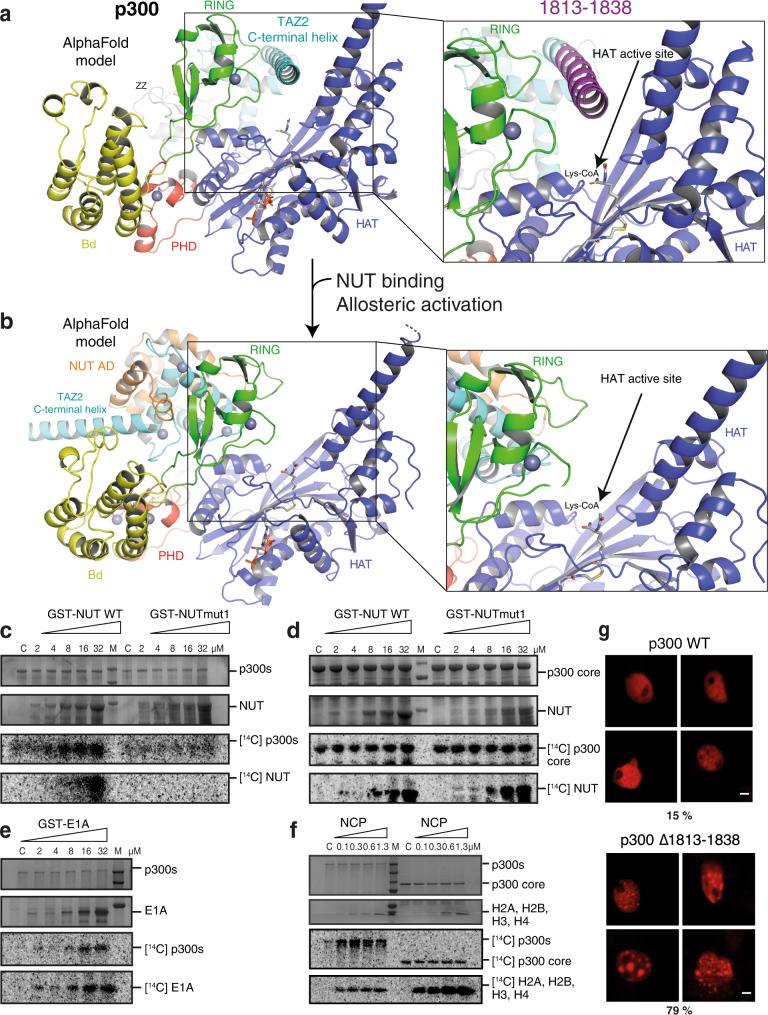
Fig. 4. Role of the TAZ2 domain in p300 regulation and substrate acetylation.
a AlphaFold prediction of p300 core-CH3 (1048-1838) structure. The structure is coloured according to domain structure. The TAZ2 domain is positioned in the HAT active site thus autoinhibiting substrate access. The TAZ2 C-terminal helix deletion (Δ1813-1838) is coloured in magenta b NUT AD binding displaces the TAZ2 domain from the HAT active site thus enabling allosteric HAT activation. c p300s or d,p300 core were incubated with [14C]Acetyl-CoA in the presence or absence (C: p300 alone) of increasing concentrations of wild type or mut1 NUT8 (396-470). e Increasing concentrations of GST-E1a we incubated with p300s. f Increasing concentrations of NCPs were incubated with p300s or p300 core. Panels c-f: Top two images: Analysis by SDS–PAGE followed by Coomassie staining. Bottom two images: 14C phosphor imaging. Experiments c-f were repeated independently three times with consistency. Representative data are shown. Source data are provided as a Source Data file. g Cos7 cells were transfected with the HA-tagged p300 WT (top) or p300 Δ1813-1838 (bottom) and analysed by anti HA immunofluorescence. Four representative nuclei are shown. The percentage of cells showing condensates (n = 100 cells) is indicated. Experiments were repeated twice with consistency. Scale bars, 5 μm.