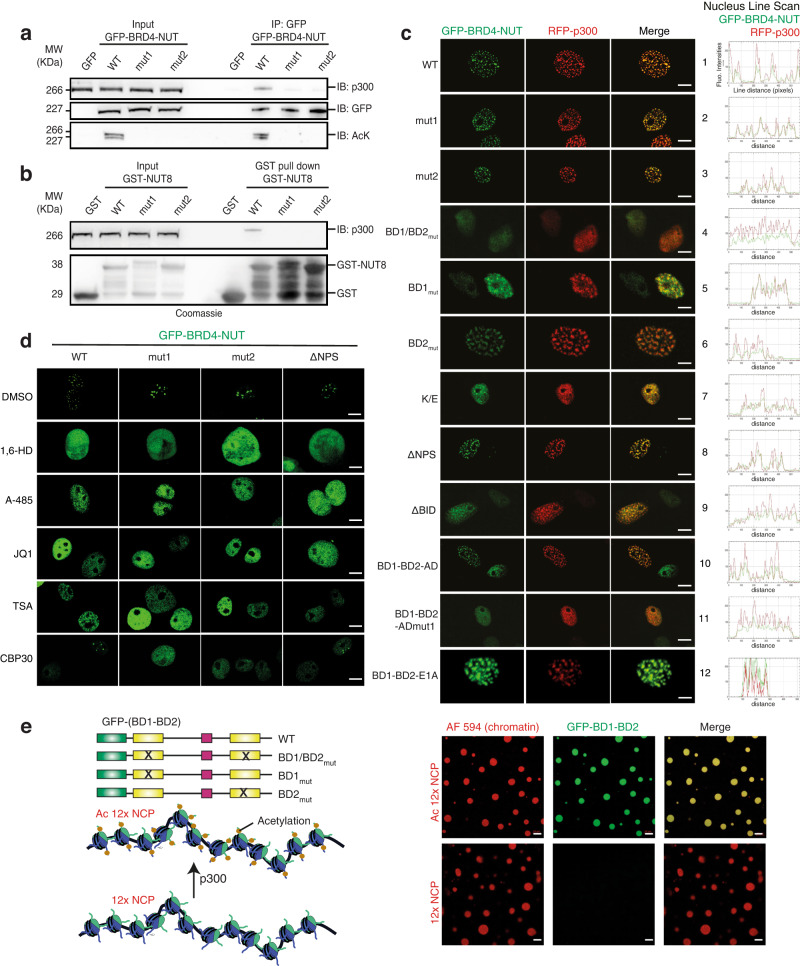
Fig. 5. Mechanism of condensation of BRD4-NUT and p300.
a Cos7 cells were transfected with GFP-BRD4-NUT variants or GFP alone as the control. Co-immunoprecipitation (IP) experiments were performed using anti-GFP antibody and immuno-blotted (IB) for p300, GFP and Acetylated-Lysine. Input samples were loaded as a control. b Purified GST-NUT8 wild type and mutants 1 and 2 were incubated with Cos7 cells extract for 4 h. The complexes were pull-down by Glutathione Sepharose beads and analysed by SDS-PAGE followed by Coomassie staining (bottom) or by IB for p300 (top). Source data are provided as a Source Data file. Experiments in a-b we repeated independently at least three times. Representative data are shown. c Cos7 cells were transfected with the indicated GFP-BRD4-NUT and RFP-p300 constructs and analysed by fluorescence microscopy, BRD4-NUT (green) and p300 (red). Scale bars, 10 μm. Quantification of foci formation of different BRD4-NUT variants and their co-localization with p300 are shown in Supplementary Tables 5, 6. d Cos7 cells were transfected with the indicated GFP-BRD4-NUT constructs and treated with DMSO (control), 1,6-hexanediol, p300 HAT inhibitor A485, pan-BET inhibitor JQ1, HDAC inhibitor TSA or p300 bromodomain inhibitor CBP30. Scale bars, 10 μm. Quantification of foci formation in presence of CBP30 is shown in Supplementary Table 6. e Schematic representation of GFP-labelled BRD4 mutants and Alexa Fluor 594 labelled reconstituted chromatin containing non-acetylated (12x NCP) or acetylated (Ac 12X NCP) 12 Nucleosome Core Particles. Fluorescence microscopy images of non-acetylated or acetylated 12x NCP chromatin incubated with GFP-BD1-BD2. Experiments in c-e were repeated independently at least three times with consistency. Scale bars, 10 μm.