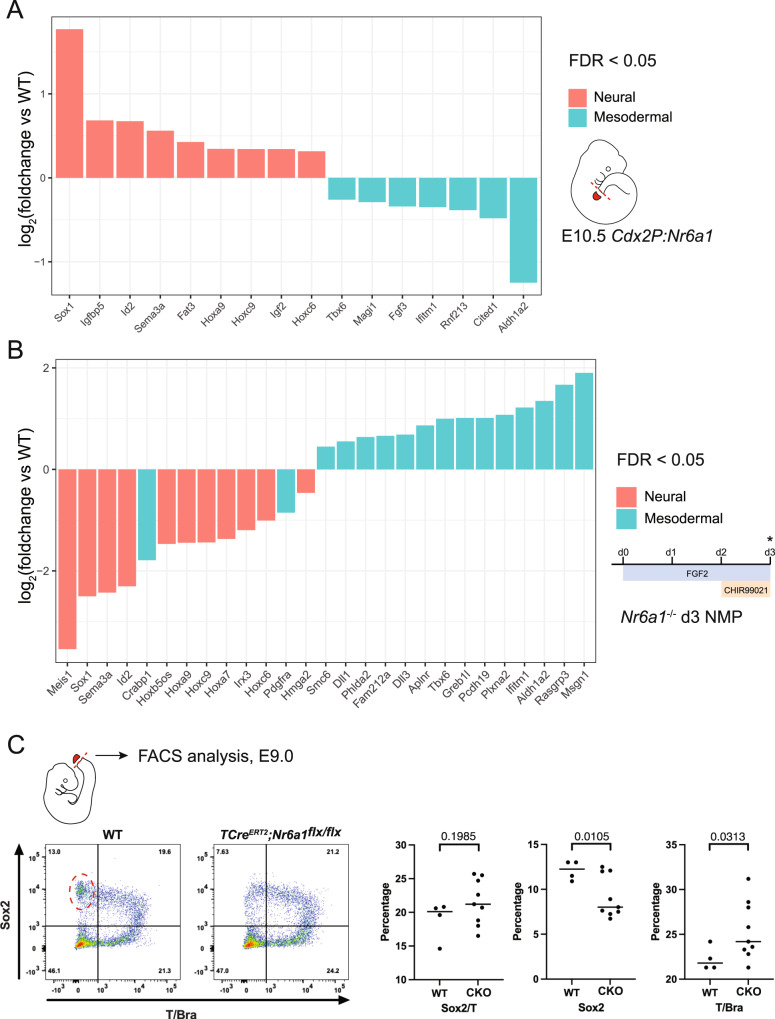
Fig. 8. Nr6a1 activity regulates neural vs mesodermal fate choice in vitro and in vivo.
A Heightened neural (red) and reduced mesodermal (blue) gene expression signatures were observed following prolonged maintenance of Nr6a1 within the mouse tailbud. RNAseq analysis was presented as a log2-transformed fold change in Cdx2P:Nr6a1 samples relative to wildtype, n = 2 for Cdx2P:Nr6a1 and n = 4 for wildtype. Neural and mesodermal gene lists were selected based on in vivo single-cell RNAseq analysis20, and only genes with a false discovery rate (FDR) < 0.05 are displayed. Source data are provided as a Source Data file. B Heightened mesodermal (blue) and reduced neural (red) gene expression signatures were observed in Nr6a1−/− in vitro-derived NMPs compared to wildtype. In vitro differentiation protocol schematised, analysis time-point (d3) marked with asterisk. RNAseq analysis is presented as a log2-transformed fold change in Nr6a1−/− samples relative to wildtype, n = 3/genotype. Neural and mesodermal gene lists as per above, and only genes with a false discovery rate (FDR) < 0.05 are displayed. Source data are provided as a Source Data file. C Flow cytometry analysis of Sox2 and T/Bra protein expression within cells of the E9.0 tailbud, collected from WT (n = 4 biologically independent samples) and TCreERT2;Nr6a1flx/flx (n = 9 biologically independent samples) embryos. Single Sox2-positive, single T/Bra-positive, or dual positive cells are presented as a percentage of all single, viable cells. Statistical comparison between genotypes was performed using an unpaired t-test with Welch’s correction, p-values are provided in the figure.