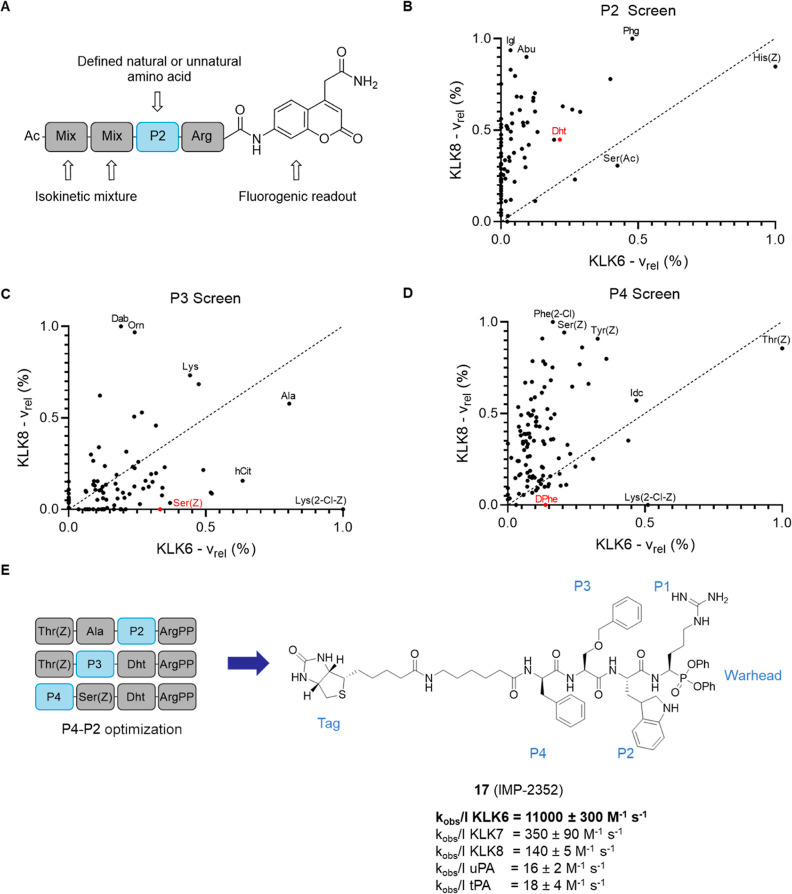
Figure 2.
Design of selective KLK ABPs using a fluorogenic peptide library. (A) Structure of peptides in the library, exemplified by the P2 sub-library. The isokinetic mixture results in an equimolar ratio of all natural amino acids at the randomized positions but with norleucine replacing methionine and cysteine. (B–D) Scatter plots comparing the substrate specificities of KLK6 and KLK8 in the P2, P3, and P4 positions. The axes represent the cleavage velocity (vrel) of a peptide in the library, relative to the highest velocity (set to 1) for KLK6 (x-axis) and KLK8 (y-axis). Amino acids above the dashed line are preferred by KLK8, while amino acids below the dashed line are preferred by KLK6. The top three amino acids for each KLK are labeled in black, while the final optimized amino acid for the KLK6 ABP is labeled in red. (E) P2–P4 probe optimization strategy. All probes were tested for potency (kobs/I) using pseudo-first-order kinetics. Structure and selectivity of optimized biotinylated KLK6 ABP 17 (IMP-2352).