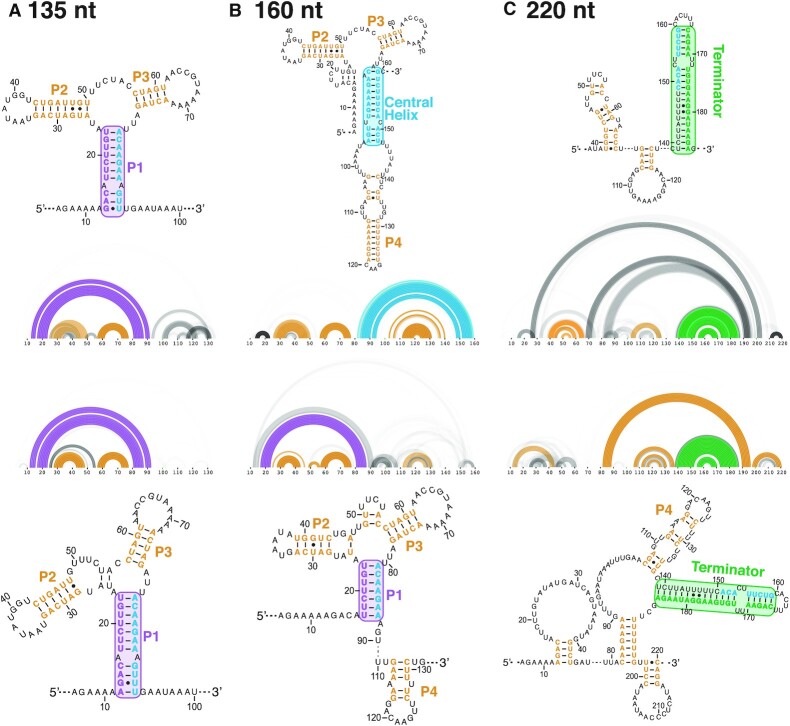
Figure 3.
Experimentally informed models of structural intermediates along the yxjA folding pathway without ligand. Cotranscriptional SHAPE-Seq experiments were performed at two intermediate lengths of (A) 135 nt, (B) 160 nt and (C) for the full-length riboswitch (220 nt) in the absence of 2AP. Reactivity data were then incorporated into R2D2 pathway modeling to generate 100 structures that are maximally consistent with the reactivity data. This family of structures is displayed as RNAbow plots, where an arc between two positions indicates a base pair in a specific structure, and the opacity of the arc indicates the prevalence of the base pair among the selected structures. Two replicates were performed at each length. The consensus structure over the population of selected structures is shown as a secondary structure alongside each bow plot. In the secondary structures, the central helix is colored teal while the 5′ side of the P1 helix is purple and the 3′ side of the terminator is green. Colored boxes drawn around the P1 helix, the central helix, or the terminator, as well as the color of the P2/P3/P4 helices, match the color of arcs in the RNAbow plots.