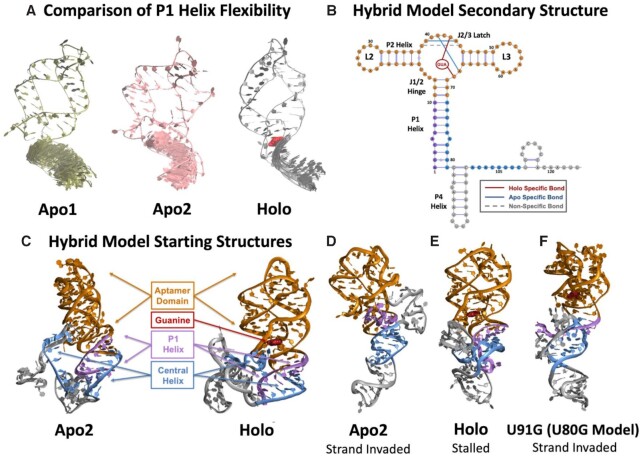
Figure 6.
Snapshots of MD and 2D REMD simulations. (A) A visual comparison of every 500th frame of the 3′-strand of the P1 helix in the apo1, apo2 and holo aptamers after a 50 ns MD simulation shows increased rigidity from the apo states to the holo state. (B) The secondary structure of the hybrid model of the yxjA riboswitch used in MD simulations. Hydrogen bonds that form in apo and holo states within the aptamer junction region are annotated, alongside interactions with the ligand. (C) Three-dimensional starting structures of the 124 nt hybrid model of the apo2 and holo states after a 50 ns equilibration. Different domains are color coded as indicated. (D) An ending conformation of one 2D REMD simulation of the apo2 hybrid model showing successful strand exchange of the P1 helix to form the central helix. (E) An ending conformation of one 2D REMD simulation of the holo hybrid model showing a stalled strand exchange process that leaves the P1 helix intact. (F) An ending conformation of one 2D REMD simulation on the broken ‘ON’ U91G mutant (U80G in the hybrid model) holo hybrid model showing successful strand exchange of the P1 helix to form the central helix.