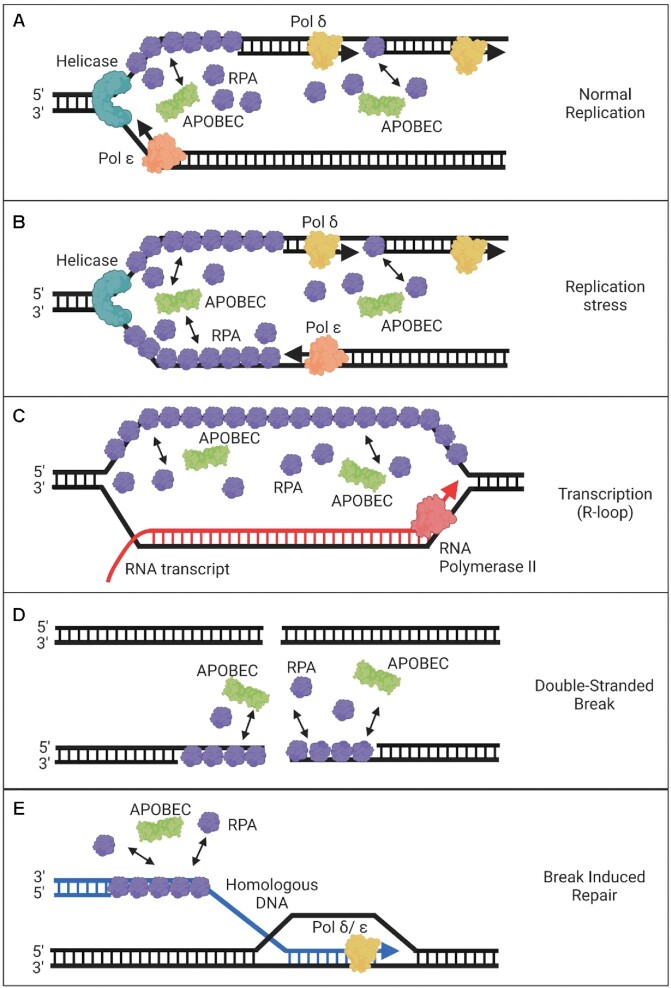
Figure 3.
Models of the RPA/APOBEC interface during DNA replication, transcription, and repair. (A) During normal replication, APOBEC enzymes compete with RPA primarily for access to the lagging strand where there is more ssDNA. (B) Replication stress can cause uncoupling of the DNA polymerase with helicase and can result in larger regions of ssDNA that are bound by RPA on both the leading and lagging strands. APOBECs can compete with the RPA that accumulates at these sites for access to ssDNA. (C) RPA can bind the non-template DNA strand of a stalled transcription bubble that has formed an R-loop. The R-loop is intentionally depicted larger than what would occur in a cell to illustrate potential protein–DNA interactions. Although there does not appear to be a major role of RPA during normal transcription, it does accumulate when transcription stalls or on R-loops. The APOBEC enzyme AID is more likely to deaminate during transcription than A3 enzymes, although some A3 activity during transcription has been demonstrated. (D) DNA repair resulting from DSB can lead to end-resection of DNA exposing ssDNA that is bound by RPA. APOBECs compete with RPA for access to this DNA, which may be facilitated if there is an RPA deficiency in the cell due to excessive firing of replication forks or high amounts of DNA damage. (E) BIR gives extended access to ssDNA. The migrating bubble replication during BIR creates ssDNA that persists and is thought to be a major source of A3-induced kataegis.