Figure 3.
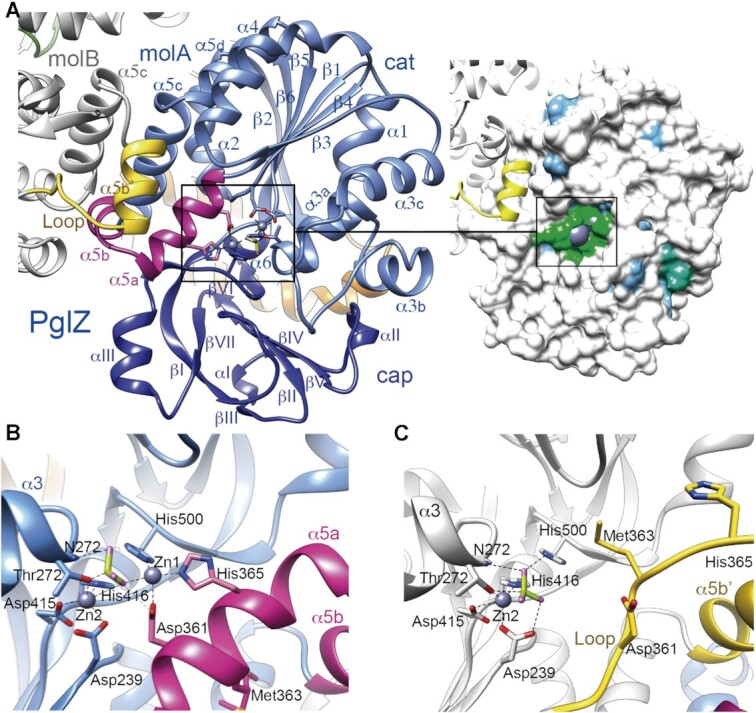
Crystal structure of the PglZ domain. (A) Left panel, Overall structure of the PglZ domain. Monomers A (molA, in blue) and B (molB, in grey) are shown. The catalytic (cat) and cap subdomains are shown in light and dark blue, respectively. Secondary structure elements are labelled. MolA helices α5a and α5b are depicted in magenta, whereas the same region in molB is folded into a loop and helix α5b′, shown in yellow. The two Zn2+ ions, BeF3 and coordinating side chains at the active site are depicted as ball and stick models. Note that without α5a–α5b, the root mean square deviation (RMSD) between molecules drops from 1.66 Å to 1.07 Å for Cα atoms 212–518. Right panel, the Connolly surface with the conservation map depicted on the PglZ domain, which shows low conservation to its five closest structural homologs (all of them are phosphodiesterases), except for the metal-binding residues. Identical amino acids are coloured in green, similar (75%) in blue and non-conserved (<50% in white). See also Supplementary Figure 2. (B) PglZ domain with helices α5a and α5b (HH conformation) in the PorX molA active site. (C) PglZ domain with the loop and helix α5b′ (LH conformation) in PorX molB. Panels (B) and (C) use the same colour code as in (A).
