Figure 3.
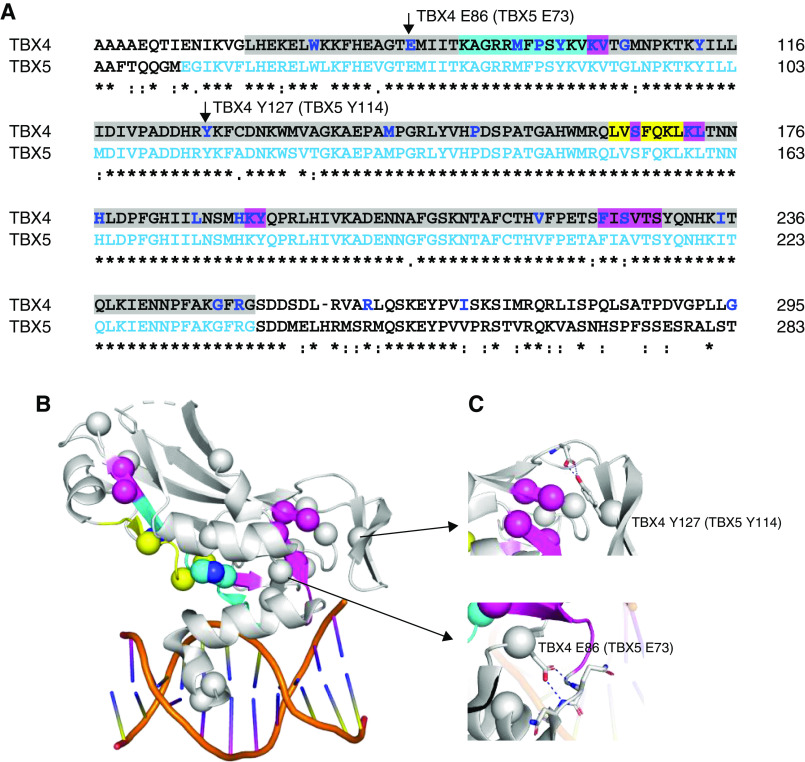
Structural analysis of TBX4 (T-BOX transcription factor 4) sequence variants. The crystal structure of TBX5 (T-BOX transcription factor 5) bound to DNA, Protein Database code 2X6V, was used for structural analysis. They share 52.6% sequence together with the full-length proteins and 93.9% in the T-BOX domain. (A) Sequence alignment of TBX4 and TBX5 in the T-BOX region (highlighted in gray) containing the DNA-binding motif as well as the nuclear localization segment 1 (NLS1, in cyan) and the nuclear export segment (NES, in yellow) (29, 30). TBX4 missense variants are indicated in bold/blue, with indels highlighted in magenta. Residues visible in the TBX5 structure are shown in light blue letters. (B) Mutations plotted on the TBX5 crystal structure as spheres. Cyan, yellow, and magenta spheres correspond to the NLS1, NES regions, and indels as indicated in A. When annotating loss-of-function variants on the TBX4 sequence, they are highly enriched in the T-BOX, particularly the NLS1 and NES. (C) Some mutations of the noninterface residues, such as TBX4 p.Glu86 and p.Tyr127 (corresponding residues p.Glu73 and p.Tyr114 in TBX5, respectively), make essential interactions to stabilize the secondary structural elements required for T-BOX binding to DNA. Clustal Omega was used for sequence alignment. Figures were generated using PyMOL Molecular Graphics System.