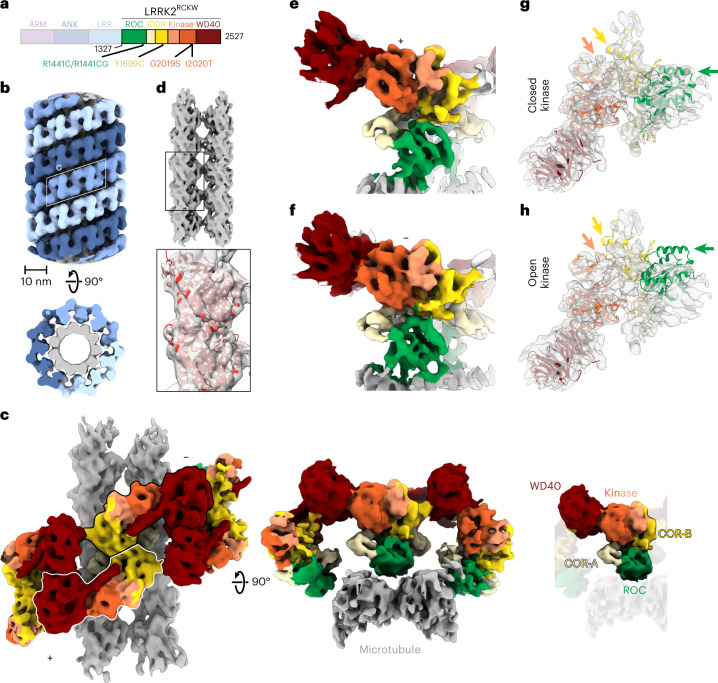
Fig. 1. Cryo-EM structure of microtubule-associated LRRK2RCKW-I2020T.
a, Primary structure of LRRK2. The N-terminal half of LRRK2, absent from the construct used in our cryo-EM studies, is shown in dim colors. The same color-coding of domains is used throughout the figures. b, Helical reconstruction (18 Å) of LRRK2RCKW-I2020T filaments bound to a microtubule in the presence of MLi-2. The three LRRK2RCKW-I2020T helices are indicated in different shades of blue. c, Cryo-EM reconstruction (6.6 Å) of a LRRK2RCKW tetramer and associated microtubule (two protofilaments), as indicated by the white rhomboid in b. Two views are shown, along with a separate representation with a single monomer highlighted and its domains labeled. d, Focused refinement of the microtubule in c to improve its resolution and determine its polarity. An α/β-tubulin dimer (from PDB: 6O2R) was docked into the density (black rectangle and inset below). e,f, Focused refinement of the ‘+’ (5.0 Å) and ‘−’ (5.2 Å) LRRK2RCKW-I2020T monomers (as labeled in c). g, The LRRK2RCKW domains (ROC, COR-A, COR-B, kinase N-lobe, kinase C-lobe, WD40) (PDB:6VNO) were fitted individually into the 4.5-Å cryo-EM map. h, The full LRRK2RCKW model (PDB: 6VNO) was aligned to the C-lobe of the kinase, as docked in g. The colored arrows in g and h point to parts of the model (PDB:6VNO) that fit the cryo-EM density better when domains are docked individually, allowing the kinase to be in a closed conformation (g), but to protrude from it when the full model is used, which has its kinase in an open conformation (h).