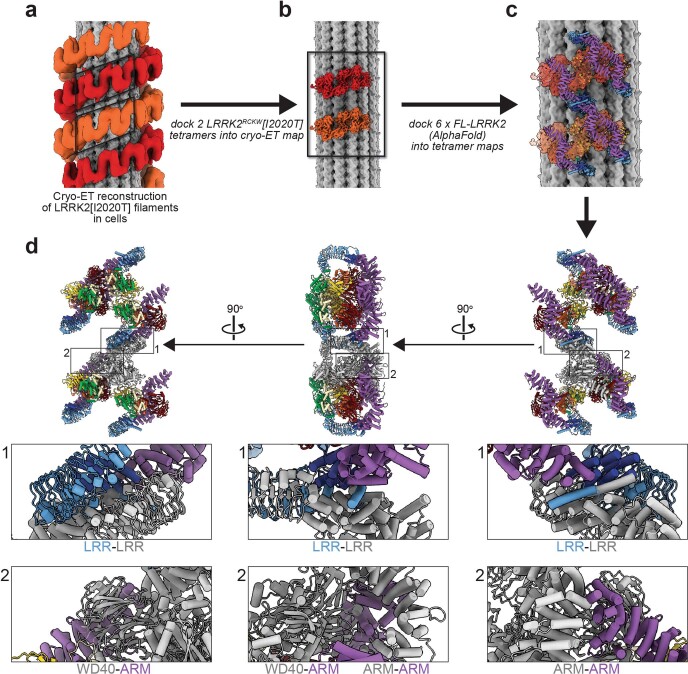
Extended Data Fig. 6. Modeling of full-length LRRK2 into the cryo-ET reconstruction of microtubule-associated LRRK2[I2020T] filaments in cells.
a, Cryo-ET reconstruction of microtubule-associated LRRK2[I2020T] filaments in cells1. The LRRK2 strands that form the double-helical filaments are shown in light and dark orange. For this figure, the density corresponding to the microtubule was replaced with a 10 Å representation of a molecular model of a microtubule. b, We docked copies of our 5.9 Å reconstruction of a LRRK2RCKW[I2020T] tetramer from the microtubule-associated filaments (Fig. 1b, c) into the regions indicated by the parallelograms in (a). c, Next, we docked two copies of the AlphaFold model of full-length LRRK2 (AF-Q5S007), which is in the active state, as is the case with LRRK2RCKW[I2020T] in our filaments, into each of the 5.9 Å maps. The pairs of AlphaFold models in each map correspond to the COR-B:COR-B dimer. This panel shows a region corresponding to the rectangle in (b). d, Three different views of the models docked in (c). Below each model, close-ups show regions where adjacent filaments clash. These clashes involve a domain in the N-terminal repeats of one LRRK2, and either the same domain on another LRRK2, or the WD40 domain. For clarity, one of the LRRK2’s is shown in grey instead of the standard rainbow coloring.