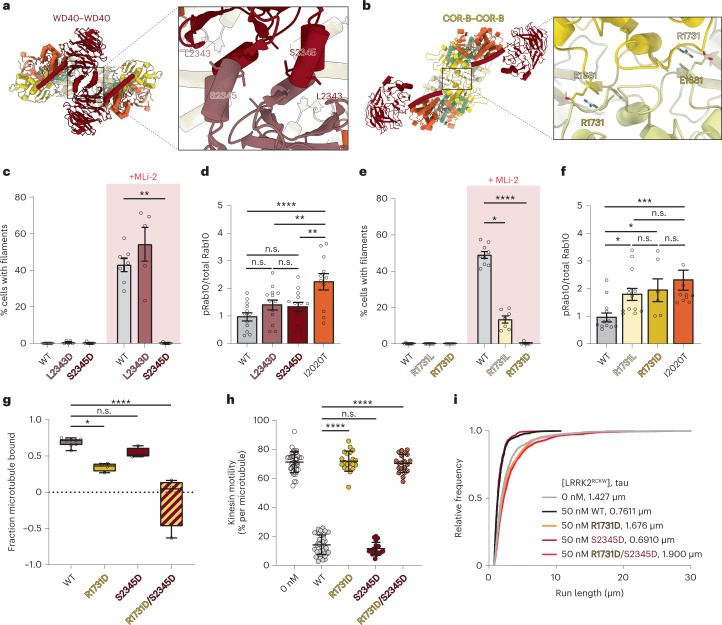
Fig. 2. Effect of mutations in LRRK2’s WD40 and COR-B domains on filament formation and microtubule binding.
a,b, Dimer interfaces (WD40–WD40 and COR-B–COR-B) involved in filament formation, and the location of the residues tested in this work. c, Effect of mutations in the WD40 domain (p.L2343D or p.S2345D) that reduce dimerization of the isolated domain in vitro or the formation of MLi-2-induced filaments in cells. Individual data points represent separate coverslips of cells obtained across at least three independent experiments. Data are mean ± s.e.m. **P = 0.0076, Kruskal–Wallis test with Dunn’s post hoc for multiple comparisons. d, Rab10 phosphorylation in 293T cells overexpressing WT LRRK2, LRRK2 carrying mutations in the WD40 domain, or LRRK2-I2020T, which increases Rab10 phosphorylation in cells. 293T cells were transiently co-transfected with plasmids encoding GFP-LRRK2 (WT or mutant) and GFP-Rab10. Quantified immunoblotting data are shown as p-Rab10/total GFP-Rab10 ratios, normalized to the average of all WT values. Individual data points represent separate populations of cells obtained across at least three independent experiments. Data are mean ± s.e.m. ****P < 0.0001, **P < 0.0052, one-way ANOVA followed by a Fisher’s least-significant difference test. e, Effect of mutations (p.R1731L or p.R1731D) at the COR-B–COR-B interface on the formation of MLi-2-induced filaments in cells. Individual data points represent separate coverslips of cells obtained from at least three independent experiments. Data are mean ± s.e.m. *P = 0.0205, ****P < 0.0001, Kruskal–Wallis test with Dunn’s post hoc for multiple comparisons. f, Rab10 phosphorylation in 293T cells overexpressing WT LRRK2, LRRK2 with mutations in the COR-B domain, or LRRK2-I2020T. 293T cells were treated as in d. Data are quantified and shown as in d. Data are mean ± s.e.m. *P < 0.035, ***P = 0.0010, one-way ANOVA followed by a Fisher’s least-significant difference test. g, Effect of mutations in the WD40 or WD40 and COR-B domains on the binding of LRRK2RCKW to microtubules in a microtubule pelleting assay. Box and whisker plot center line denotes the median value; whiskers denote the minimum and maximum values. *P = 0.0111, ****P < 0.0001, one-way ANOVA with Dunnett’s multiple comparisons test. WT, n = 8 replicates; mutants, n = 4 replicates. h, Effect of mutations in the WD40 or WD40 and COR-B domains on the inhibition of kinesin motility in vitro by 50 nM LRRK2RCKW. Inhibition of kinesin motility was quantified as percentage of motile events per microtubule. Data points represent individual microtubules obtained across at least two independent experiments. Data are mean ± s.d. ****P < 0.0001, Kruskal–Wallis test with Dunn’s post hoc for multiple comparisons. i, Cumulative distribution of run lengths for kinesin in the absence or presence of 50 nM LRRK2RCKW (WT or carrying WD40, COR-B, or WD40 and COR-B mutations). The run lengths were not significantly different between 50 nM WT and LRRK2RCKW-S2345D, and were significantly different between 50 nM WT LRRK2RCKW and LRRK2RCKW-R1731D S2345D and LRRK2RCKW-R1731D (Kruskal–Wallis test with Dunn’s post hoc for multiple comparisons). Mean decay constants (tau) are shown.