Abstract
Background
Infection during pregnancy can increase the risk of neurodevelopmental disorders in offspring. The impact of maternal SARS-CoV-2 infection on infant neurodevelopment is poorly understood. The maternal immune response to infection may be mimicked in rodent models of maternal immune activation which recapitulate altered neurodevelopment and behavioural disturbances in the offspring. In these models, epigenetic mechanisms, in particular DNA methylation, are one pathway through which this risk is conferred in utero to offspring. We hypothesised that in utero exposure to SARS-CoV-2 in humans may alter infant DNA methylation, particularly in genes associated with neurodevelopment. We aimed to test this hypothesis in a pilot sample of children in Victoria, Australia, who were exposed in utero to SARS-CoV-2.
Methods
DNA was extracted from buccal swab specimens from (n = 4) SARS-CoV-2 in utero exposed and (n = 4) non-exposed infants and methylation status assessed across 850,000 methylation sites using an Illumina EPIC BeadChip. We also conducted an exploratory enrichment analysis using Gene Ontology annotations.
Results
1962 hypermethylated CpG sites were identified with an unadjusted p-value of 0.05, where 1133 CpGs mapped to 959 unique protein coding genes, and 716 hypomethylated CpG sites mapped to 559 unique protein coding genes in SARS-CoV-2 exposed infants compared to non-exposed. One differentially methylated position (cg06758191), located in the gene body of AFAP1 that was hypomethylated in the SARS-CoV-2 exposed cohort was significant after correction for multiple testing (FDR-adjusted p-value <0.00083). Two significant differentially methylated regions were identified; a hypomethylated intergenic region located in chromosome 6p proximal to the genes ZP57 and HLA-F (fwer <0.004), and a hypomethylated region in the promoter and body of the gene GAREM2 (fwer <0.036). Gene network enrichment analysis revealed differential methylation in genes corresponding to pathways relevant to neurodevelopment, including the ERBB pathway.
Conclusion
These pilot data suggest that exposure to SARS-CoV-2 in utero differentially alters methylation of genes in pathways that play a role in human neurodevelopment.
Keywords: SARS-CoV-2, Maternal, Neurodevelopment, In utero, DNA methylation
Highlights
-
•
SARS-CoV-2 exposure in utero alters infant DNA methylation.
-
•
The gene AFAP1 is hypomethylated in SARS-CoV-2 exposed infants.
-
•
SARS-CoV-2 exposed infants show hypomethylation of a region in the gene GAREM2.
-
•
SARS-CoV-2 exposed infants show enriched methylation in genes belonging to neurodevelopmental pathways.
1. Introduction
The COVID-19 pandemic has created unprecedented acute global health challenges but the long-term impacts of COVID-19 infection, particularly during prenatal development are yet to be understood. Although transplacental or vertical transmission of SARS-CoV-2 has been reported, it appears to be rare (Hosier et al., 2020); however, of considerable concern is the maternal immune response to SARS-CoV-2 and the so called ‘cytokine storm’ that is a common occurrence following infection. Concern over this immune response is borne from previous ecological studies, birth cohort studies and animal models that have established key links between the activation of pro-inflammatory pathways in the mother with adverse neurodevelopmental outcomes in the infant (Atladottir et al., 2010; Brown, 2012; Meyer et al., 2005).
Mouse model studies have shown that one potential mechanism for how maternal immune activation (MIA) could alter offspring brain development is via DNA methylation changes (Basil et al., 2014; Labouesse et al., 2015; Richetto et al., 2017). Multiple clinical studies have reported associations between maternal adversities, such as stress, gestational diabetes and obesity, with altered infant DNA methylation (Godfrey et al., 2017; Howe et al., 2020; Tian et al., 2019). In addition, a growing body of research points to abnormal DNA methylation early in life as a potential causative factor for neurodevelopmental disorders such as Autism Spectrum Disorder (ASD) (Tremblay and Jiang, 2019) and schizophrenia (Hannon et al., 2021). However, no studies that we are aware of have assessed DNA methylation following MIA in human infants. Given the potential for SARS-CoV-2 infection to induce a significant immune response, we hypothesised that infants exposed in utero to maternal SARS-CoV-2 may show altered DNA methylation compared to non-exposed, age and sociodemographic matched controls. With the recent finding that SARS-CoV-2 exposure in utero increases the risk of neurodevelopmental disorder in offspring (Edlow et al., 2022), we hypothesise that differential methylation of genes and/or groups of genes in defined pathways involved in neurodevelopment occurs in infants exposed in utero to SARS-CoV-2.
2. Materials and methods
2.1. Ethics approval and consent
The study was approved through Monash Health Human Research Ethics Committee RES-20-0000-801A (protocol #6, 17/03/2022). Adult participants provided signed consent prior to participation in the study for themselves and their child. The work described was carried out in accordance with The Code of Ethics of the World Medical Association for experiments involving humans.
2.1.1. Participant recruitment
A total of n = 8 mother-infant dyads (4 SARS-CoV-2 exposed and 4 non-exposed) were recruited from Monash Health (Melbourne, Australia) between January to July 2021. Pregnant women who presented at a Monash Health site with a positive SARS-CoV-2 PCR test were enrolled into our ongoing prospective cohort study. Controls were matched as closely as possible to SARS-CoV-2 exposed women by month of delivery, gestational age of their child at birth and socio-demographic status. Women who had a history of substance dependence, family violence or psychiatric illness were excluded. Maternal demographic data and past medical and obstetric history were collected through face-to-face interview. The WHO-7 point ordinal scale (Organization, 2020) was used to rank the severity of infection for women who tested positive to SARS-CoV-2 during pregnancy. The Edinburgh Postnatal Depression Scale (EPND) (Cox et al., 1987) and Maternal Postnatal Attachment Scale (MPAS) (Condon and Corkindale, 1998) were administered to all mothers. Child weight, length and head circumference were recorded and the Hammersmith Infant Neurological Examination objective assessment was performed at 3 months by a trained health professional. This study is part of a larger longitudinal study that includes ongoing neurodevelopmental assessments for the child from birth to 15 years of age (Hill et al., 2022).
2.1.2. Biospecimen collection and storage
Buccal swabs from all infants were collected at ∼3 months of age and stored at −20 °C until extraction. Samples were de-identified at collection and allocated a study code.
2.1.3. DNA extraction and bisulphite conversion
DNA was extracted from buccal swab samples using the Isohelix Buccal-Prep Plus DNA Isolation kit according to manufacturer protocol (Cell Projects, Kent). Sample purity was assessed using a Nanodrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and DNA concentration quantified using a Qubit fluorometer (Thermo Scientific, Wilmington, DE). Bisulphite conversion of 500 ng input DNA was performed according to manufacturers’ instructions using the Zymo EZ DNA Methylation kit (Zymo Research, Irvine, CA). Methylation profiling was conducted using the Infinium MethylationEPIC 8 sample BeadChip (Illumina, San Diego, CA) and imaged using the Illumina iScan System (Illumina, San Diego, CA).
2.1.4. Genome-wide DNA methylation analysis
Raw signal intensity data (.idat) files were imported into RStudio (version 1.4.1717) and data quality checking followed a pre-processing pipeline provided by the R package “ChAMP” (Morris et al., 2014). Probes with a detection p-value >0.01 in at least 50% of samples were excluded. Probes located in known SNPs, aligning to multiple locations, non-CpG probes and probes on X and Y chromosomes were also excluded from further analysis. Beta-mixture quantile normalisation was performed to correct for type II probe bias (Teschendorff et al., 2013).
2.1.5. Identification of differentially methylated CpG sites and regions
R package “limma” (Smyth, 2004; Wettenhall and Smyth, 2004) was used to identify differentially methylated CpG sites in SARS-CoV-2 exposed samples (N = 4) compared to non-exposed (N = 4). Genomic regions were annotated using HumanMethylationEPIC probe annotations using the R package “ChAMP” (Morris et al., 2014). To reduce type I errors, raw p-values were corrected for probe-wise multiple testing using the Benjamini-Hochberg (BH) method to produce FDR-corrected q-values. The Bumphunter method (Jaffe et al., 2012) was employed to further assess for differentially methylated regions (DMRs) in SARS-CoV-2 exposed samples (N = 4) compared to non-exposed (N = 4), using all nominally significant probes to determine segments within the genome which displayed consistently hyper or hypomethylated CpGs (Peters et al., 2015).
2.1.6. Exploratory pathway analyses
Differentially methylated sites with a log fold-change (logFC) greater than 0.1 or less than −0.1 and a nominal p-value of less than 0.05 were submitted to further downstream analysis. The resulting differentially methylated sites were annotated to 1518 genes, upon which gene-set enrichment analysis (using the ORA method calculated based on hypergeometric distribution) was performed using Gene Ontology (GO) annotations with “clusterprofiler” and “pathview” packages (Wu et al., 2021; Yu et al., 2012).
3. Results/discussion
3.1. The characteristics of the study population are shown in Table 1
Table 1.
Descriptive statistics of the study cohort.
| Maternal Factors | SARS-CoV-2 Exposed (n = 4) | Controls(n = 4) | P-value |
|---|---|---|---|
| Age (years), median (SD) | 27.5 (3.59) | 28.5 (4.08) | 0.66 |
| Number of previous pregnancies, median (SD) | 1 (1.15) | 0 (1.00) | 0.54 |
| Highest educational attainment, n(%) | |||
| High school or less | 0 | 1(25) | |
| Diploma/TAFE | 1(25) | 2(50) | |
| University degree or higher | 3(75) | 1(25) | |
| Employment status, n(%) | |||
| Not working or on maternity leave | 2(50) | 0 | |
| Part-time | 1(25) | 2(50) | |
| Full-time | 1(25) | 2(50) | |
| Language spoken at home, n(%) | |||
| English | 1(25) | 4(100) | |
| Other | 3(75) | 0 | |
| Marital status, n(%) | |||
| Married | 4(100) | 4(100) | |
| Single | 0 | 0 | |
| Recent immigrant ( < 5 years), n(%) | |||
| Yes | 3(75) | 0 | |
| No | 1(25) | 4(100) | |
| Pregnancy complications, n(%) | |||
| Gestational diabetes | 2(50) | 0 | |
| FGR | 1(25) | 0 | |
| Hypertensive disorders of pregnancy | 0 | 2(50) | |
| PROM | 0 | 1(20) | |
| Mode of delivery, n(%) | |||
| Normal vaginal delivery | 3(75) | 2(50) | |
| Caesarean section | 1(25) | 2(50) | |
| Gestation of infection, n(%) | |||
| 2nd trimester | 1(25) | ||
| 3rd trimester | 3(75) | ||
| WHO 7-point Ordinal Scale, n(%) | |||
| 1 (Ambulatory, no limitation of activities) | 3(75) | ||
| 4 (Hospitalised, low-flow supplemental O2) | 1(25) | ||
| Edinburgh Postnatal Depression Score, mean (SD) | 4.5(3.11) | 3.25(2.50) | 0.55 |
| Maternal Postnatal Attachment Score, mean (SD) | 84.98(6.29) | 86.83(5.92) | 0.68 |
| Child Factors | |||
|---|---|---|---|
| Sex, n(%) | |||
| Female | 0 | 1(25) | |
| Male | 4(100) | 3(75) | |
| Gestational age at birth n(%) | |||
| <37 weeks | 1(25) | 2(50) | |
| >/37 weeks | 3(75) | 2(50) | |
| Birth weight (median, kg ± SD) | 3.32 ± 0.76 | 3.65 ± 0.43 | 0.37 |
| Birth length (median, cm ± SD) | 49 ± 4.34 | 51.5 ± 2.94 | 0.73 |
| Birth head circumference (median, cm ± SD) | 34 ± 3.3 | 36 ± 2.09 | 0.69 |
| Hammersmith Infant Neurological Exam score (3 months) | 66.5 ± 5.4 | 63.3 ± 2.7 | 0.62 |
Abbreviations - FGR: fetal growth restriction, O2: oxygen, PROM: premature rupture of membranes. Weight, length and head circumference data are raw median values standard deviation (SD) and p-values were generated using a paired t-test.
Of the four participants who contracted COVID-19 during pregnancy, three participants scored a 1 in the WHO 7-point ordinal scale for COVID-19 illness severity. This score means they had very mild symptoms with no limitations of activities. One mother scored a 4 on the WHO ordinal scale, meaning she was hospitalised and on low-flow supplemental oxygen. No participants were on any medications and no participants were vaccinated as all participants were infectious in late 2020, prior to any approved medication or vaccinations. Only one mother was potentially infectious at the time of labour (tested positive 3 days before delivery), however, she scored quite low for illness severity (1) and gave birth by normal vaginal delivery. Two mothers in the SARS-CoV-2 exposed group, but none in the control group had gestational diabetes.
3.1.1. Methylation profiling of protein-coding genes in SARS-CoV-2 exposed vs control infants
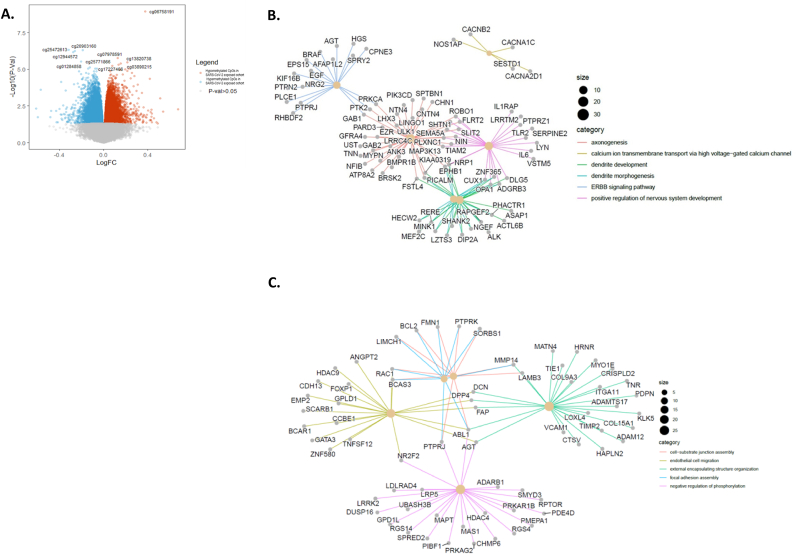
Of the differentially methylated CpG sites with a logFC greater than 0.1 or less than −0.1 and an unadjusted p-value of 0.05, we found 1962 hypermethylated CpG sites, where 1133 CpGs mapped to 959 unique protein coding genes, and 716 hypomethylated CpG sites mapped to 559 unique protein coding genes using the illumine annotation (Fig. 1A). One site corresponding to probe cg06758191 and located in the gene body of AFAP1 (CHR 4: 7812988) was hypomethylated (logFC = 0.373760432) in the SARS-CoV-2 exposed cohort and remained significant after correction for multiple testing (q-value <0.00083).
Fig. 1.
– (A) Volcano plot of differential methylation analysis with the x axis displaying log fold-change (log fc) and the y axis displaying the log10 of p values for each CpG site. CpGs with relative hypermethylation (- log fc) in SARS-CoV-2 exposed cohort and relative hypomethylation ( + log fc) are represented by blue and red dots respectively. Grey dots represent CpGs below the FDR = 0.05 threshold. Probe cg06758191, corresponding to the gene AFAP1 remained significant after multiple correction. (p < 1.13e-09, log fc = 0.3736)
(B) Gene concept network maps depicting the linkages of genes that contained CpG sites which were hypermethylated in the SARS-CoV-2 exposed cohort and (C) hypomethylated in the SARS-CoV-2 exposed cohort and their biological concepts deriving from annotated GO terms as a network. The legend on the right depicts the central categories or GO terms grouping the various genes. Genes at the junction of different colour coded lines are those that play a role across multiple biological concepts or GO terms. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Actin filament-associated protein (AFAP1) acts as an adaptor protein and regulates actin cytoskeletal organisation and integrity (Dorfleutner et al., 2007; Qian et al., 2000). AFAP1 has been identified as a key regulator of the TNFα-induced reduction in P-glycoprotein efflux function in human brain capillary endothelial cells (Hoshi et al., 2017). The P-glycoprotein efflux transporter is located at the apical surface of brain capillary epithelial cells that form the blood-brain barrier and acts to prevent or restrict blood-brain barrier entry (Blanchette and Daneman, 2015). While AFAP1 has multiple peripheral roles in addition to its role at the blood brain barrier, it would be of interest to further investigate, using animal models, if prenatal exposure to SARS-CoV-2 alters AFAP1 expression in the CNS and blood brain barrier function.
3.1.2. SARS-CoV-2 exposed infants showed differential methylated regions in chromosome 2 and 6
Two differentially methylated regions were statistically significant; one was located in chromosome 6p proximal to the genes ZP57 and HLA-F (family-wise error rate (fwer) < 0.004) (Supplementary Table 1). This region includes the major histocompatibility complex and is associated with multiple risk loci for schizophrenia and bipolar disorder, including rs6932590 and rs3131296 (Williams et al., 2011). A second region in chromosome 2 in the promoter and body regions of the gene GAREM2 (fwer <0.036) had significantly lower levels of methylation in the SARS-CoV-2 exposed cohort (Supplementary Table 1). The majority of the CpGs measured in this DMR are located in the promoter region, and all CpGs are also located within a CpG island or shore. GAREM2 is a downstream adaptor of the epidermal growth factor receptor (EGFR), which has been shown to interact with scaffolding proteins associated with the reelin pathway, and regulate neuronal differentiation and neurite outgrowth in neuroblastoma cells in mice (Taniguchi et al., 2013).
3.1.3. Gene concept network analysis showed enrichment in neurodevelopmental pathways
Gene concept network maps depicting the linkages of genes that contained CpG sites which were hypermethylated (Fig. 1B) in the SARS-CoV-2 exposed infants show hypermethylation of “axonogenesis”, “calcium ion transport”, “dendrite development”, dendrite morphogenesis”, “positive regulation of nervous system development” and “ERBB signalling pathway”. In general, genes with hypermethylation within their promotor region show inhibited or silenced expression, however recent studies have shown that there is a subset of genes whereby hypermethylation was associated with increased expression (Rauluseviciute et al., 2020) and this is dependent on the sites of methylation in a locus (Dhar et al., 2021). In terms of the hypermethylated pathways identified in our study, genes identified from the ERBB family, such as EGF, show a negative correlation of reduced mRNA expression with DNA methylation (Li et al., 2014). This is also the case for genes involved in “dendrite morphogenesis”, such as SHANK2 (Fu et al., 2020) and ALK (Gomez et al., 2015) and “axogenesis” and “positive regulation of nervous system development” such as the Slit/Robo genes (Zheng et al., 2009).
ERBB signalling pathway enrichment is of particular note as we have previously shown altered ERBB (epidermal growth factor receptor) and reelin expression in the brains of mice exposed to MIA in utero that were accompanied by disrupted cognitive constructs relevant to schizophrenia, including working memory and cognitive flexibility (Nakamura et al., 2022). EGF pathway dysfunction is also apparent in schizophrenia, with post-mortem studies showing reduced EGF but increased EGFR expression in the prefrontal cortex of people with schizophrenia (Futamura et al., 2002).
Hypomethylated gene networks in SARS-CoV-2 exposed infants include “cell-substrate junction assembly”, “endothelial cell migration”, “external encapsulating structure organisation”, “focal adhesion assembly” and “negative regulation of phosphorylation networks” (Fig. 1C). Hypomethylation or loss of DNA methylation is generally associated with increased gene expression (Ehrlich, 2009). This association has previously been noted for genes involved in “endothelial cell migration”, such as FOXP1 (Garaud et al., 2017) and “negative regulation of phosphorylation networks”, such as MAPT (Coupland et al., 2014).
Cell-cell adhesions play a crucial role in synapse formation, plasticity, and cell signalling (O'Dushlaine et al., 2011). Meanwhile altered serine/threonine kinase activity and disrupted phosphorylation has been suggested to play a role in the pathophysiology of schizophrenia (Chadha et al., 2021).
4. Conclusion
While considerable caution must be taken when extrapolating these findings as many of these pathways are not specific to the CNS, these data suggest that exposure to SARS-CoV-2 in utero differentially alters methylation of several gene pathways that play a role in neurodevelopment. Limitations of this study include the small sample size and the lack of ability to associate any peripheral DNA methylation findings to alterations in the brain. However, using the tool IMAGE-CpG (Braun et al., 2019)(http://han-lab.org/methylation/default/imageCpG), accessed on 03/11/2022), which is a data base of methylation changes in 13 individuals with matched samples of brain, buccal, blood and saliva, we found that brain DNA methylation of the gene AFAP1 correlates with buccal AFAP DNA methylation to a relatively high degree (r2 = 0.74, p < 0.0001). While the above correlation is encouraging the authors concede that with a small N of 4 cases and 4 controls, this data set should be considered as a pilot project with preliminary, albeit provocative, findings. Here, further replication with larger numbers, as well as animal model studies to confirm that peripheral DNA methylation associates with brain transcriptome changes, are required to substantiate these identified pathways.
Furthermore, we are unable to determine if any methylation changes found in our study are specific to SARS-CoV-2, or may be more generally attributed to infections and the maternal immune response, as previously suggested (Brown, 2012; Brown et al., 2004; Mednick et al., 1988). Within the context of this small study we are unable to differentiate this, however, rodent maternal immune activation modelling data, which shows gene expression changes in the same ERBB and GABAergic pathways that were differentially methylated in the SARS-CoV-2 exposed infants, certainly suggest that the maternal immune response may be the common mediating factor causing altered gene expression of particular pathways (Nakamura et al., 2022). While all women in this study were not vaccinated as they were pregnant prior to vaccine availability, findings could potentially be different in vaccinated women, who we anticipate will have a milder course of illness and immune response. Finally, a significant consideration when extrapolating these data is that it was not possible to perfectly match cases and controls or to consider the use of covariates, due to the small samples size. Here, an important health related issue that could not be controlled for was that two of the SARS-CoV-2 exposed mothers but none of the non-exposed mothers had gestational diabetes during pregnancy, which may impact infant DNA methylation (Howe et al., 2020). Planned longitudinal prospective monitoring of the infants neurodevelopment (Hill et al., 2022) with larger sample sizes will enable identification of whether methylation changes observed correlate with neurodevelopmental disorders or other maternal factors captured from this valuable cohort.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the philanthropic organisation, One in Five.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2022.100572.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Atladottir H.O., Thorsen P., Schendel D.E., Ostergaard L., Lemcke S., Parner E.T. Association of hospitalization for infection in childhood with diagnosis of autism spectrum disorders: a Danish cohort study. Arch. Pediatr. Adolesc. Med. 2010;164:470–477. doi: 10.1001/archpediatrics.2010.9. [DOI] [PubMed] [Google Scholar]
- Basil P., Li Q., Dempster E.L., Mill J., Sham P.C., Wong C.C., McAlonan G.M. Prenatal maternal immune activation causes epigenetic differences in adolescent mouse brain. Transl. Psychiatry. 2014;4 doi: 10.1038/tp.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Daneman R. Formation and maintenance of the BBB. Mech. Dev. 2015;138 Pt 1:8–16. doi: 10.1016/j.mod.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Braun P., Han S., Nagahama Y., Gaul L., Heinzman J., Hing B., Grossbach A., Dlouhy B., Howard M., Kawasaki H., Potash J., Shinozaki G. 28 - IMAGE-CpG: development of a web-based search tool for genome-wide DNA methylation correlation between live human brain and peripheral tissues within individuals. Eur. Neuropsychopharmacol. 2019;29:S796. doi: 10.1038/s41398-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S. Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Developmental neurobiology. 2012;72:1272–1276. doi: 10.1002/dneu.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S., Begg M.D., Gravenstein S., Schaefer C.A., Wyatt R.J., Bresnahan M., Babulas V.P., Susser E.S. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch. Gen. Psychiatr. 2004;61:774–780. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- Chadha R., Alganem K., McCullumsmith R.E., Meador-Woodruff J.H. mTOR kinase activity disrupts a phosphorylation signaling network in schizophrenia brain. Mol. Psychiatr. 2021;26:6868–6879. doi: 10.1038/s41380-021-01135-9. [DOI] [PubMed] [Google Scholar]
- Condon J.T., Corkindale C.J. The assessment of parent-to-infant attachment: development of a self-report questionnaire instrument. J. Reprod. Infant Psychol. 1998;16:57–76. [Google Scholar]
- Coupland K.G., Mellick G.D., Silburn P.A., Mather K., Armstrong N.J., Sachdev P.S., Brodaty H., Huang Y., Halliday G.M., Hallupp M., Kim W.S., Dobson-Stone C., Kwok J.B. DNA methylation of the MAPT gene in Parkinson's disease cohorts and modulation by vitamin E in vitro. Mov. Disord. : Off. J. Mov. Disord. Soc. 2014;29:1606–1614. doi: 10.1002/mds.25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.L., Holden J.M., Sagovsky R. Detection of postnatal depression. Development of the 10-item edinburgh postnatal depression scale. Br. J. Psychiatr. : J. Ment. Sci. 1987;150:782–786. doi: 10.1192/bjp.150.6.782. [DOI] [PubMed] [Google Scholar]
- Dhar G.A., Saha S., Mitra P., Nag Chaudhuri R. DNA methylation and regulation of gene expression: guardian of our health. Nucleus (Calcutta) 2021;64:259–270. doi: 10.1007/s13237-021-00367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfleutner A., Stehlik C., Zhang J., Gallick G.E., Flynn D.C. AFAP-110 is required for actin stress fiber formation and cell adhesion in MDA-MB-231 breast cancer cells. J. Cell. Physiol. 2007;213:740–749. doi: 10.1002/jcp.21143. [DOI] [PubMed] [Google Scholar]
- Edlow A.G., Castro V.M., Shook L.L., Kaimal A.J., Perlis R.H. Neurodevelopmental outcomes at 1 Year in infants of mothers who tested positive for SARS-CoV-2 during pregnancy. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Liu D., Guo J., Long H., Xiao W., Xiao W., Feng L., Luo Z., Xiao B. Dynamic change of shanks gene mRNA expression and DNA methylation in epileptic rat model and human patients. Mol. Neurobiol. 2020;57:3712–3726. doi: 10.1007/s12035-020-01968-5. [DOI] [PubMed] [Google Scholar]
- Futamura T., Toyooka K., Iritani S., Niizato K., Nakamura R., Tsuchiya K., Someya T., Kakita A., Takahashi H., Nawa H. Abnormal expression of epidermal growth factor and its receptor in the forebrain and serum of schizophrenic patients. Mol. Psychiatr. 2002;7:673–682. doi: 10.1038/sj.mp.4001081. [DOI] [PubMed] [Google Scholar]
- Garaud S., Roufosse F., De Silva P., Gu-Trantien C., Lodewyckx J.N., Duvillier H., Dedeurwaerder S., Bizet M., Defrance M., Fuks F., Bex F., Willard-Gallo K. FOXP1 is a regulator of quiescence in healthy human CD4(+) T cells and is constitutively repressed in T cells from patients with lymphoproliferative disorders. Eur. J. Immunol. 2017;47:168–179. doi: 10.1002/eji.201646373. [DOI] [PubMed] [Google Scholar]
- Godfrey K.M., Reynolds R.M., Prescott S.L., Nyirenda M., Jaddoe V.W., Eriksson J.G., Broekman B.F. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017;5:53–64. doi: 10.1016/S2213-8587(16)30107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez S., Castellano G., Mayol G., Sunol M., Queiros A., Bibikova M., Nazor K.L., Loring J.F., Lemos I., Rodriguez E., de Torres C., Mora J., Martin-Subero J.I., Lavarino C. DNA methylation fingerprint of neuroblastoma reveals new biological and clinical insights. Epigenomics. 2015;7:1137–1153. doi: 10.2217/epi.15.49. [DOI] [PubMed] [Google Scholar]
- Hannon E., Dempster E.L., Mansell G., Burrage J., Bass N., Bohlken M.M., Corvin A., Curtis C.J., Dempster D., Di Forti M., Dinan T.G., Donohoe G., Gaughran F., Gill M., Gillespie A., Gunasinghe C., Hulshoff H.E., Hultman C.M., Johansson V., Kahn R.S., Kaprio J., Kenis G., Kowalec K., MacCabe J., McDonald C., McQuillin A., Morris D.W., Murphy K.C., Mustard C.J., Nenadic I., O'Donovan M.C., Quattrone D., Richards A.L., Rutten B.P., St Clair D., Therman S., Toulopoulou T., Van Os J., Waddington J.L., Wellcome Trust Case Control, C., consortium, C. Sullivan P., Vassos E., Breen G., Collier D.A., Murray R.M., Schalkwyk L.S., Mill J. DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia. Elife. 2021;10 doi: 10.7554/eLife.58430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R., Malhotra A., Sackett V., Williams K., Fahey M., Palmer K.R., Hunt R.W., Darke H., Lim I., Newman-Morris V., Cheong J.L.Y., Whitehead C., Said J., Bignardi P., Muraguchi E., Fernandes L.C.C., Jr., Oliveira C., Sundram S. A prospective, longitudinal, case-control study protocol to evaluate the neurodevelopment of children from birth to adolescence exposed to COVID-19 in utero. Protoc. Exch. 2022 doi: 10.21203/rs.3.pex-1980/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y., Uchida Y., Tachikawa M., Ohtsuki S., Terasaki T. Actin filament-associated protein 1 (AFAP-1) is a key mediator in inflammatory signaling-induced rapid attenuation of intrinsic P-gp function in human brain capillary endothelial cells. J. Neurochem. 2017;141:247–262. doi: 10.1111/jnc.13960. [DOI] [PubMed] [Google Scholar]
- Hosier H., Farhadian S.F., Morotti R.A., Deshmukh U., Lu-Culligan A., Campbell K.H., Yasumoto Y., Vogels C.B., Casanovas-Massana A., Vijayakumar P., Geng B., Odio C.D., Fournier J., Brito A.F., Fauver J.R., Liu F., Alpert T., Tal R., Szigeti-Buck K., Perincheri S., Larsen C., Gariepy A.M., Aguilar G., Fardelmann K.L., Harigopal M., Taylor H.S., Pettker C.M., Wyllie A.L., Cruz C.D., Ring A.M., Grubaugh N.D., Ko A.I., Horvath T.L., Iwasaki A., Reddy U.M., Lipkind H.S. SARS-CoV-2 infection of the placenta. J. Clin. Invest. 2020;130:4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe C.G., Cox B., Fore R., Jungius J., Kvist T., Lent S., Miles H.E., Salas L.A., Rifas-Shiman S., Starling A.P., Yousefi P., Ladd-Acosta C., Baccarelli A., Binder E.B., Chatzi V.L., Czamara D., Dabelea D., DeMeo D.L., Ghantous A., Herceg Z., Kajantie E., Lahti J.M.T., Lawlor D.A., Litonjua A., Nawrot T.S., Nohr E.A., Oken E., Pizzi C., Plusquin M., Raikkonen K., Relton C.L., Sharp G.C., Sorensen T.I.A., Sunyer J., Vrijheid M., Zhang W., Hivert M.F., Breton C.V. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the pregnancy and childhood epigenetics consortium. Diabetes Care. 2020;43:98–105. doi: 10.2337/dc19-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A.E., Murakami P., Lee H., Leek J.T., Fallin M.D., Feinberg A.P., Irizarry R.A. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 2012;41:200–209. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M.A., Dong E., Grayson D.R., Guidotti A., Meyer U. Maternal immune activation induces GAD1 and GAD2 promoter remodeling in the offspring prefrontal cortex. Epigenetics. 2015;10:1143–1155. doi: 10.1080/15592294.2015.1114202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Fan J., Li Z., Xu C. DNA methylation dynamics in the rat EGF gene promoter after partial hepatectomy. Genet. Mol. Biol. 2014;37:439–443. doi: 10.1590/s1415-47572014000300017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mednick S.A., Machon R.A., Huttunen M.O., Bonett D. Adult schizophrenia following prenatal exposure to an influenza epidemic. Arch. Gen. Psychiatr. 1988;45:189–192. doi: 10.1001/archpsyc.1988.01800260109013. [DOI] [PubMed] [Google Scholar]
- Meyer U., Feldon J., Schedlowski M., Yee B.K. Towards an immuno-precipitated neurodevelopmental animal model of schizophrenia. Neurosci. Biobehav. Rev. 2005;29:913–947. doi: 10.1016/j.neubiorev.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Morris T.J., Butcher L.M., Feber A., Teschendorff A.E., Chakravarthy A.R., Wojdacz T.K., Beck S. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30:428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J.P., Schroeder A., Gibbons A., Sundram S., Hill R.A. Timing of maternal immune activation and sex influence schizophrenia-relevant cognitive constructs and neuregulin and GABAergic pathways. Brain Behav. Immun. 2022;100:70–82. doi: 10.1016/j.bbi.2021.11.006. [DOI] [PubMed] [Google Scholar]
- O'Dushlaine C., Kenny E., Heron E., Donohoe G., Gill M., Morris D., International Schizophrenia C., Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol. Psychiatr. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- Organization W.H. 2020. WHO R&D Blueprint Novel Coronavirus COVID-19 Therapeutic Trial Synopsis. [Google Scholar]
- Peters T.J., Buckley M.J., Statham A.L., Pidsley R., Samaras K., V Lord R., Clark S.J., Molloy P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Baisden J.M., Zot H.G., Van Winkle W.B., Flynn D.C. The carboxy terminus of AFAP-110 modulates direct interactions with actin filaments and regulates its ability to alter actin filament integrity and induce lamellipodia formation. Exp. Cell Res. 2000;255:102–113. doi: 10.1006/excr.1999.4795. [DOI] [PubMed] [Google Scholar]
- Rauluseviciute I., Drablos F., Rye M.B. DNA hypermethylation associated with upregulated gene expression in prostate cancer demonstrates the diversity of epigenetic regulation. BMC Med. Genom. 2020;13:6. doi: 10.1186/s12920-020-0657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richetto J., Massart R., Weber-Stadlbauer U., Szyf M., Riva M.A., Meyer U. Genome-wide DNA methylation changes in a mouse model of infection-mediated neurodevelopmental disorders. Biol. Psychiatr. 2017;81:265–276. doi: 10.1016/j.biopsych.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Tanaka S., Ishii A., Watanabe M., Fujitani N., Sugeo A., Gotoh S., Ohta T., Hiyoshi M., Matsuzaki H., Sakai N., Konishi H. A brain-specific Grb2-associated regulator of extracellular signal-regulated kinase (Erk)/mitogen-activated protein kinase (MAPK) (GAREM) subtype, GAREM2, contributes to neurite outgrowth of neuroblastoma cells by regulating Erk signaling. J. Biol. Chem. 2013;288:29934–29942. doi: 10.1074/jbc.M113.492520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F.Y., Rifas-Shiman S.L., Cardenas A., Baccarelli A.A., DeMeo D.L., Litonjua A.A., Rich-Edwards J.W., Gillman M.W., Oken E., Hivert M.F. Maternal corticotropin-releasing hormone is associated with LEP DNA methylation at birth and in childhood: an epigenome-wide study in Project Viva. Int. J. Obes. 2019;43:1244–1255. doi: 10.1038/s41366-018-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M.W., Jiang Y.H. DNA methylation and susceptibility to autism spectrum disorder. Annu. Rev. Med. 2019;70:151–166. doi: 10.1146/annurev-med-120417-091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Williams H.J., Craddock N., Russo G., Hamshere M.L., Moskvina V., Dwyer S., Smith R.L., Green E., Grozeva D., Holmans P., Owen M.J., O'Donovan M.C. Most genome-wide significant susceptibility loci for schizophrenia and bipolar disorder reported to date cross-traditional diagnostic boundaries. Hum. Mol. Genet. 2011;20:387–391. doi: 10.1093/hmg/ddq471. [DOI] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Liu B.B., Liu Y.K., Kang X.N., Sun L., Guo K., Sun R.X., Chen J., Zhao Y. [Analysis of the expression of Slit/Robo genes and the methylation status of their promoters in the hepatocellular carcinoma cell lines] Zhonghua Gan Zang Bing Za Zhi. 2009;17:198–202. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.