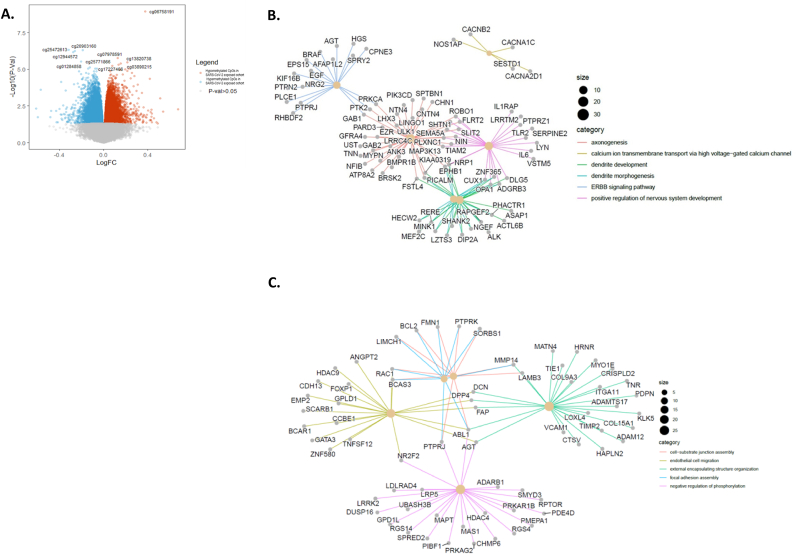
Fig. 1.
– (A) Volcano plot of differential methylation analysis with the x axis displaying log fold-change (log fc) and the y axis displaying the log10 of p values for each CpG site. CpGs with relative hypermethylation (- log fc) in SARS-CoV-2 exposed cohort and relative hypomethylation ( + log fc) are represented by blue and red dots respectively. Grey dots represent CpGs below the FDR = 0.05 threshold. Probe cg06758191, corresponding to the gene AFAP1 remained significant after multiple correction. (p < 1.13e-09, log fc = 0.3736)
(B) Gene concept network maps depicting the linkages of genes that contained CpG sites which were hypermethylated in the SARS-CoV-2 exposed cohort and (C) hypomethylated in the SARS-CoV-2 exposed cohort and their biological concepts deriving from annotated GO terms as a network. The legend on the right depicts the central categories or GO terms grouping the various genes. Genes at the junction of different colour coded lines are those that play a role across multiple biological concepts or GO terms. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)