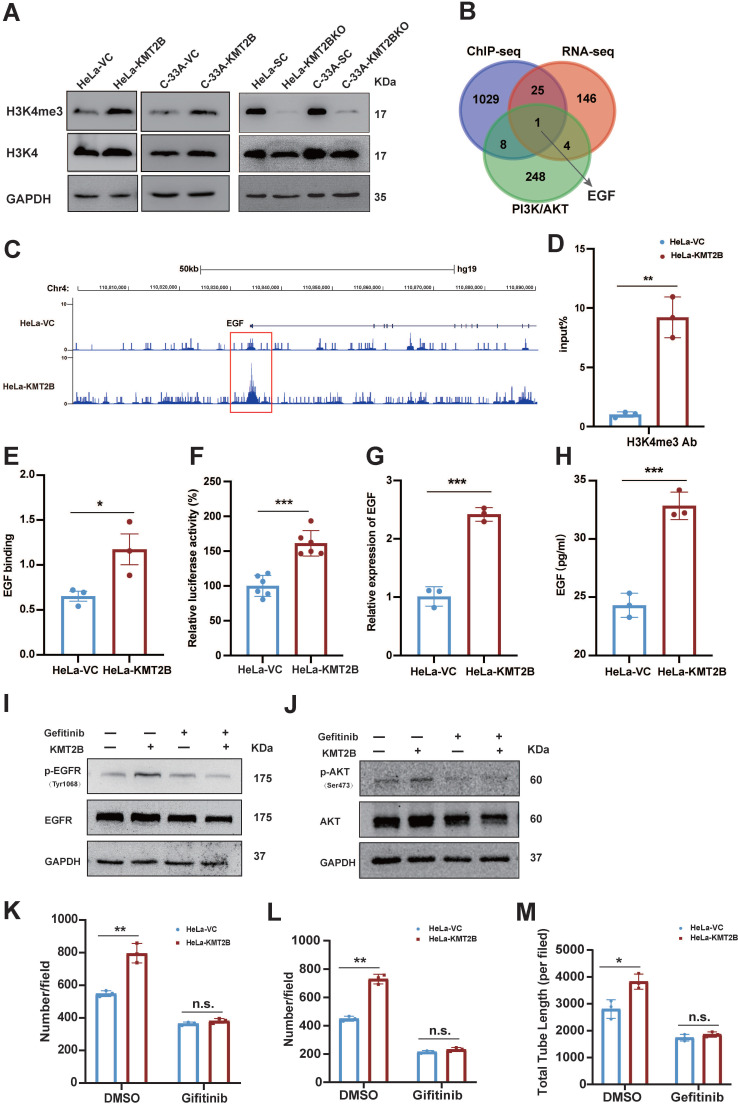
Figure 7.
EGF is a target gene of KMT2B in CC cells and mediates KMT2B-dependent PI3K/AKT activity. (A) Effects of KMT2B overexpressing on the global levels of histone H3K4 tri-methylation using western blotting analysis. (B) Intersection analysis of differential genes in RNA-seq, CHIP-seq and PI3K/AKT signaling pathway related genes. (C) Visualization of the H3K4me3 enrichment in the promoter region of EGF gene HeLa-VC and HeLa-KMT2B cells. (D) H3K4me3 CHIP-qPCR analysis was used to determine H3K4me3 level at EGF promoter region in HeLa-VC and HeLa-KMT2B cells. (E) ChIP-qPCR was used to verify the binding of KMT2B at the EGF promoter region in HeLa-VC and HeLa-KMT2B cells. (F) Luciferase reporter genes driven by the EGF promoter (pGL3-EGF) were co-transfected with pRL-TK into HeLa-VC and HeLa-KMT2B cells, and luciferase signals were measured after 48 h. The relative luciferase activity value in HeLa-VC cells was set to 100%. (F) qPCR assays confirmed the difference in expression of EGF gene between HeLa-VC and HeLa-KMT2B cells at mRNA level. (G) ELISA assays confirmed the difference in expression of EGF gene between HeLa-VC and HeLa-KMT2B cells at protein level. (H, I) Western blotting analysis was used to detect the expression of p-EGFR and p-AKT after treatment of gefitinib in HeLa-VC and HeLa-KMT2B cells. (J) HeLa-VC and HeLa-KMT2B cells with or without gefitinib treatment were cultured in non-coated chambers for 8 h. The cell migration was quantitated by counting the migrated cells. (K) HeLa-VC and HeLa-KMT2B cells with or without gefitinib treatment were cultured in Matrigel-coated chambers for 12 h. The cell invasion was quantitated by counting the invaded cells. (L) Quantitated the Total length of tubes by Image Pro Plus software after incubating in HeLa-VC and HeLa-KMT2B condition medium with or without gefitinib treatment for 4 h. Data represents mean ± SD (n = 3). All the experiments have been repeated at least once with similar results. *, p < 0.05, **, p < 0.01; ***, p < 0.001, ns, not significant.