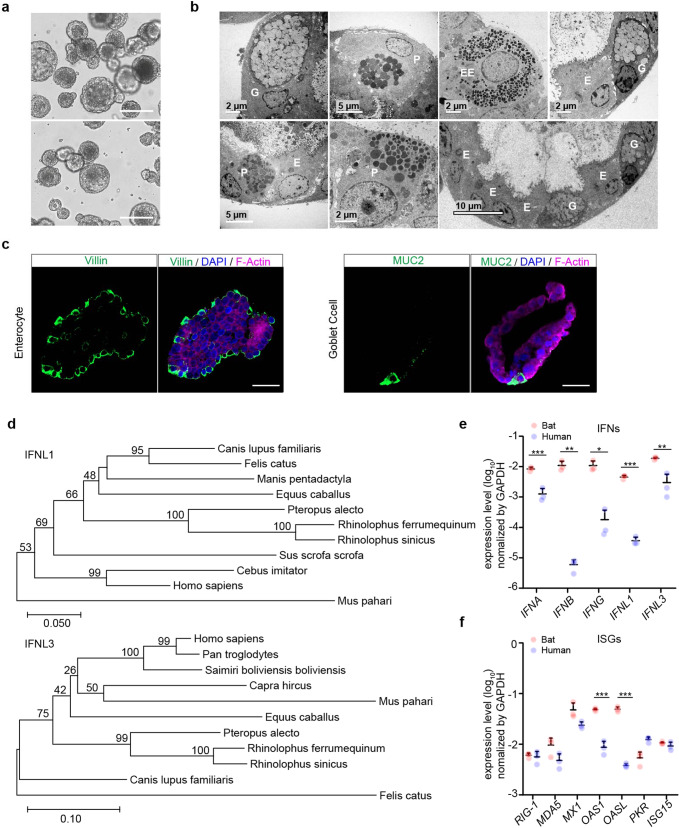
Fig. 1.
Characterization of optimized bat intestinal organoids and detection of the basal expression of antiviral genes. a Photomicrographs of optimized bat intestinal organoids on day 7 after passaging (magnification ×100). Scale bar, 100 μm. b Transmission electron microscopy illustrates the ultrastructural morphology of absorptive enterocyte (E), Paneath cell (P), goblet cell (G), and enteroendocrine cell (EE) in bat intestinal organoids. c Bat intestinal organoids were fixed and immunostained to label Villin + (green) enterocyte and MUC2 + (green) goblet cell. Nuclei and actin filaments were counterstained with DAPI (blue) and Phalloidin-647 (purple), respectively. Scale bar, 20 µm. d Phylogenetic analysis based on an alignment of horseshoe bat (Rhinolophus sinicus) IFNL1 and IFNL3 ORF cDNAs with other Chiroptera species and mammals. Bootstrap values (%) are indicated on each branch, and the scale for branch length is shown at the bottom of the tree. e, f GAPDH-normalized expression levels of IFN genes (e) and ISGs (f) in bat and human intestinal organoids. Data represent the mean and s.d. of a representative experiment in organoids from a bat and human donor, n = 3. Two-tailed unpaired Student’s t test