Abstract
Escherichia coli DNA ligase (LigA) is the prototype of the NAD+-dependent class of DNA ligases found in all bacteria. Here we report the characterization of E.coli LigB, a second NAD+-dependent DNA ligase identified by virtue of its sequence similarity to LigA. LigB differs from LigA in that it lacks the BRCA1 C-terminus domain (BRCT) and two of the four Zn-binding cysteines that are present in LigA and all other bacterial NAD+ ligases. We found that recombinant LigB catalyzed strand joining on a singly-nicked DNA in the presence of a divalent cation and NAD+, and that LigB reacted with NAD+ to form a covalent ligase-adenylate intermediate. Alanine substitution for the motif I lysine (126KxDG) abolished nick joining and ligase-adenylate formation by LigB, thus confirming that the ligase and adenylyltransferase activities are intrinsic to the LigB protein.
INTRODUCTION
NAD+-dependent DNA ligase was first isolated from Escherichia coli in 1967 (1–4) and the E.coli enzyme has since served as the prototype for mechanistic studies of the bacterial ligase family (5). Escherichia coli DNA ligase (the product of the ligA gene) catalyzes the sealing of 5′ phosphate and 3′ hydroxyl termini at nicks in duplex DNA by means of three sequential nucleotidyl transfer reactions. In the first step, attack on the adenylyl α phosphorus of NAD+ by ligase results in release of nicotinamide mononucleotide and formation of a covalent intermediate (ligase-adenylate) in which AMP is linked through a phosphoamide bond to lysine. In the second step, the AMP is transferred to the 5′ phosphate at the nick to form DNA-adenylate. In the third step, the unadenylated ligase catalyzes attack by the 3′ OH of the nick on DNA-adenylate to join the two polynucleotides and release AMP.
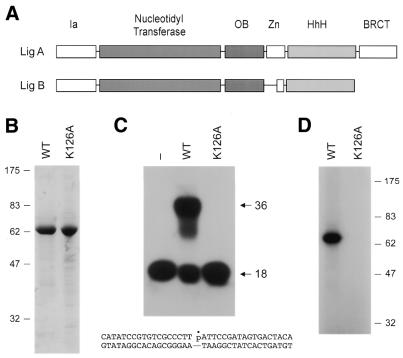
An NAD+-dependent DNA ligase is found in every bacterial species. NAD+-dependent ligases are of fairly uniform size (656–837 amino acids) and extensive amino acid sequence conservation occurs throughout the entire lengths of the polypeptides. The atomic structures of NAD+-dependent ligases from Bacillus stearothermophilus and Thermus filiformis have been determined by X-ray crystallography (6,7). Although there is scant amino acid sequence similarity between NAD+-dependent and ATP-dependent ligases, the tertiary structures of the catalytic cores are conserved and the adenylate binding pocket of NAD+ ligases is composed of the five nucleotidyl transferase motifs noted originally in the ATP-dependent enzymes (8–10). The catalytic core of the bacterial NAD+ ligase consists of nucleotidyl transferase and oligomer-binding fold (OB-fold) domains. The core is flanked by a short N-terminal domain (Ia) and three C-terminal domains: a tetracysteine domain that binds a single Zn atom; a helix–hairpin–helix domain (HhH) and a BRCT domain (named after the C-terminus of the breast cancer gene product BRCA1) (Fig. 1A).
Figure 1.
Purification and activity of E.coli LigB. (A) Comparison of the domain structures of LigA and LigB. See text for details. (B) Purification. Aliquots (4 µg) of the phosphocellulose preparations of wild-type (WT) LigB and the K126A mutant were analyzed by SDS–PAGE. Polypeptides were visualized by staining the gel with Coomassie brilliant blue dye. A photograph of the stained gel is shown. The positions and sizes (in kDa) of marker proteins are indicated on the left. (C) DNA ligation. Reaction mixtures (20 µl) containing 50 mM Tris–HCl (pH 7.5), 10 mM (NH4)2SO4, 5 mM dithiothreitol (DTT), 5 mM MgCl2, 50 µM NAD+, 1 pmol of 32P-labeled nicked DNA, and 8 pmol of wild-type (WT) LigB or K126A were incubated for 10 min at 37°C. The reactions were quenched with formamide and EDTA. The reaction products were resolved by electrophoresis through a 12% polyacrylamide gel containing 7 M urea in 90 mM Tris-borate, 2.5 mM EDTA. An autoradiogram of the gel is shown. The positions of the input 32P-labeled 18mer strand and the 36mer ligated strand are indicated by arrows on the right. The nicked duplex substrate used in the ligation reactions is illustrated at the bottom. (D) Ligase-adenylate formation. Reaction mixtures (20 µl) containing 50 mM Tris–HCl (pH 7.5), 10 mM (NH4)2SO4, 5 mM DTT, 5 mM MgCl2, 2 µM [32P-AMP]NAD+ (New England Nuclear) and 10 pmol of wild-type (WT) LigB or K126A were incubated for 5 min at 37°C. Reactions were quenched by adding SDS to 1%. The reaction products were resolved by SDS–PAGE. An autoradiogram of the dried gel is shown. The positions and sizes (in kDa) of marker proteins are indicated on the right.
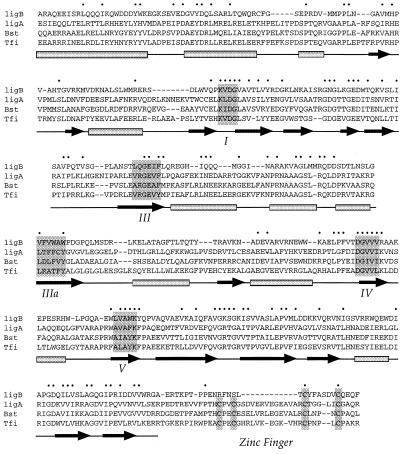
Early genetic studies showed that the ligA gene is essential for growth of E.coli (11,12). Genes encoding NAD+-dependent DNA ligase are also essential in Salmonella typhimurium, Bacillus subtilis and Staphylococcus aureus (13–15). Post-genomic analysis and biochemical studies have revealed that many bacteria contain one or more ATP-dependent DNA ligases in addition to an NAD+-dependent enzyme (16,17). This discovery raises the prospect that bacteria, like eukaryotes, may exploit different DNA ligase isozymes for different physiological functions (e.g. replication, repair, homologous recombination and non-homologous end-joining). Escherichia coli, however, does not encode an ATP-dependent ligase. Rather, a search of the complete E.coli K12 genome with the LigA protein immediately highlights a second DNA ligase-like protein (Fig. 2), encoded by an open reading frame upstream of the gene for guanylate kinase (gmk) (GenBank accession no. AE000442).
Figure 2.
Aligned primary structures of NAD+-dependent DNA ligases. The amino acid sequence of E.coli LigB from amino acids 30 to 415 is aligned with the N-terminal portions of the LigA enzymes of E.coli (Eco), B.stearothermophilus (Bst) and T.filiformis (Tfi). The alignment encompasses the Ia, nucleotidyltransferase, OB-fold and Zn finger domains. The secondary structure of Tfi ligase is shown below the amino acid sequence. Gaps in the sequence alignment are indicated by dashes. Positions of side-chain conservation (identity or structural similarity) in all four proteins are denoted by dots above the sequence. The conserved nucleotidyl transferase motifs are denoted below the Tfi sequence; motifs I, III, IIIa, IV and V are highlighted in shaded boxes. The four cysteines comprising the Zn finger are located near the C-terminus of the alignment and are highlighted in shaded boxes.
The E.coli ligase-like protein (henceforth named LigB) is a 562 amino acid polypeptide. A protein of the same size with 97% sequence identity to LigB is encoded by the enterohemorrhagic E.coli strain O157:H7 (GenBank accession no. AE005592). A manual alignment of the LigB amino acid sequence to E.coli LigA and to the Tfi and Bst ligases (guided by the crystal structure of the Tfi ligase) reveals conservation of domain Ia, the nucleotidyl transferase domain (including the five catalytic motifs) and the OB-fold (Fig. 2) as well as the HhH domain (not shown). However, the LigB protein lacks the C-terminal BRCT domain and two of the four Zn-binding cysteines that are present in all other bacterial NAD+ ligases. Given that individual cysteines of the Zn finger have been shown to be essential for the nick-joining activity of bacterial ligases (18,19), and the hypothesis that the BRCT domain plays an important role in DNA binding (20), it is of interest to evaluate whether LigB is a DNA ligase and whether it uses NAD+ as a cofactor. Here we show that this is indeed the case, although LigB is considerably less active in nick joining than is LigA.
MATERIALS AND METHODS
Expression vectors for LigB
Oligodeoxyribonucleotide primers complementary to the 5′ and 3′ ends of the E.coli K12 ligB gene were used to amplify by PCR the equivalent open reading frame from E.coli DH5α. The primers were designed to introduce NdeI and BamHI restriction sites at the 5′ and 3′ ends of the ligB gene. The PCR product was digested with NdeI and BamHI, then cloned into the NdeI and BamHI sites of pET16b (Novagen) to yield pET-LigB. Dideoxy sequencing of the entire insert of pET-LigB confirmed that no alterations of the DNA sequence were introduced during PCR amplification and cloning of the ligB gene. The K126A mutation was introduced into the ligB gene using the two stage PCR-based overlap extension method (21). An NdeI–BamHI restriction fragment of the mutated second stage PCR product was inserted into pET16b. The insert of the resulting pET-LigB-K126A plasmid was sequenced to confirm the presence of the desired alanine mutation and the absence of any unwanted coding changes.
Purification of LigB
The pET-LigB expression plasmid was transformed into E.coli BL21(DE3). A single ampicillin-resistant colony was inoculated into LB medium containing 0.1 mg/ml ampicillin and a 100 ml culture was grown at 37°C until the A600 reached 0.8. The culture was placed on ice for 30 min, then adjusted to 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and subsequently incubated at 17°C for 16 h with continuous shaking. Cells were harvested by centrifugation and the pellets were stored at –80°C. All subsequent procedures were performed at 4°C. Cell lysis was achieved by treatment of thawed, resuspended cells with 0.2 mg/ml lysozyme and 0.1% Triton X-100 in 10 ml of lysis buffer containing 50 mM Tris–HCl (pH 7.5), 0.5 M NaCl and 10% sucrose. The lysate was sonicated to reduce viscosity and insoluble material was removed by centrifugation at 37 000 g for 20 min. The supernatant was mixed with 1 ml of Ni-NTA-agarose resin (Qiagen) for 30 min with constant rotation. The slurry was poured into columns and the packed resin was washed with IMAC buffer [50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 10% glycerol] containing 5 mM imidazole. The column was step-eluted with 50 and 500 mM imidazole in IMAC buffer. The polypeptide compositions of the column fractions were monitored by SDS–PAGE. The His-tagged LigB was recovered in the 500 mM imidazole eluate (0.9 mg of protein). The preparation was diluted 1:1 with buffer P [50 mM Tris–HCl (pH 7.5), 10% glycerol] and applied to a 1 ml phosphocellulose column that had been equilibrated with buffer P. The column was washed with 200 mM NaCl in buffer P and LigB was step-eluted with 600 mM NaCl in buffer P containing 1 mM EDTA. The phosphocellulose ligase preparation (0.6 mg of protein) was stored at –80°C. The protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The K126A mutant was produced and purified from a 100 ml bacterial culture using the same protocol.
RESULTS
Purification of E.coli LigB and demonstration of ligase activity
The ligB gene was cloned into a T7 RNA polymerase-based bacterial expression vector so as to fuse the 562 amino acid LigB protein to a 20 amino acid N-terminal leader peptide containing 10 tandem histidines. The expression plasmid was introduced into E.coli BL21(DE3), a strain that contains the T7 RNA polymerase gene under the control of a lacUV5 promoter. The recombinant His-tagged protein was purified from a soluble extract of IPTG-induced bacteria by nickel-agarose affinity chromatography and phosphocellulose cation exchange chromatography steps. SDS–PAGE analysis showed that the phosphocellulose preparation was highly enriched with respect to the 64 kDa LigB polypeptide (Fig. 1B).
We assayed the ability of the recombinant LigB protein to seal a duplex DNA substrate containing a single nick (Fig. 1C). NAD+ and magnesium were included in the assay mixtures. Activity was evinced by conversion of the 5′ 32P-labeled 18mer substrate to 36mer product (Fig. 1C). Thus LigB is indeed a DNA ligase.
The initial step in DNA ligation involves formation of a covalent enzyme-adenylate intermediate. In order to assay adenylyltransferase activity, we incubated the recombinant LigB protein with [32P-AMP]NAD+ and magnesium. This resulted in the formation of a 32P-labeled covalent nucleotidyl-protein adduct that comigrated with the LigB polypeptide during SDS–PAGE (Fig. 1D, WT). We conclude that LigB is active in covalent nucleotidyl transfer with NAD+ as the AMP donor.
Alanine mutation of the motif I lysine of LigB
The KxDG sequence (motif I) is the signature feature of the ligase/capping enzyme superfamily of nucleotidyl transferases that form a covalent lysyl-NMP intermediate (8). To gauge the contribution of the motif I lysine residue (Lys126) to the activity of LigB we replaced it with alanine. The mutant protein K126A was isolated from soluble lysates by nickel-affinity and phosphocellulose chromatography; its purity was comparable to that of wild-type LigB (Fig. 1B). K126A was inert in nick ligation (Fig. 1C) and in ligase-AMP formation with [32P-AMP]NAD+ (Fig. 1D). This result is consistent with Lys126 being the site of covalent AMP attachment, as it is in other polynucleotide ligases, and it excludes the possibility that the observed nick-joining activity of the recombinant LigB protein might be attributable to contamination by the endogenous LigA.
Characterization of LigB
Initial experiments were performed to determine the optimal conditions for nick joining. LigB was active in Tris–HCl buffer from pH 6.5 to 9.0 (not shown). Nick joining by LigB depended on a divalent cation cofactor; this requirement was satisfied by either magnesium (1–10 mM) or manganese (1–10 mM) (data not shown). A comparison of various divalent cations at 5 mM concentration showed that neither calcium nor zinc supported LigB activity and that cobalt was about one-quarter as effective as magnesium (not shown).
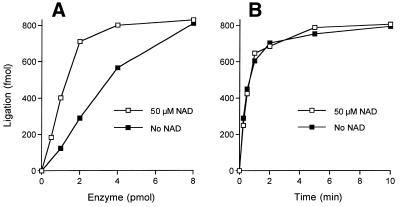
The LigB dependence of nick joining is shown in Figure 3A. Activity in the absence of added NAD+ was attributed to pre-adenylated LigB in the enzyme preparation. The linear dependence of nucleotide-independent strand joining on input enzyme suggested that 14% of the enzyme molecules had AMP bound at the active site. Inclusion of NAD+ in the reaction mixture stimulated nick ligation 3-fold. Inclusion of ATP failed to stimulate the joining reaction above the level achieved in the absence of added nucleotide. Indeed, none of the eight standard rNTPs or dNTPs were able to stimulate LigB (not shown). NADP was also inactive (not shown). NAD+ titration experiments showed that nick-joining activity was equivalent from 7 to 60 µM NAD+ and that higher NAD+ concentrations (>100 µM) were inhibitory. At ≥0.5 mM NAD+ LigB activity was reduced to the basal level seen in the absence of added nucleotide. Inhibition by 0.5–1 mM NAD+ was also observed for E.coli LigA (not shown).
Figure 3.
Ligation of nicked DNA. (A) Enzyme-dependence. Reaction mixtures (20 µl) containing 50 mM Tris–HCl (pH 7.5), 10 mM (NH4)2SO4, 5 mM DTT, 5 mM MgCl2, 1 pmol of 32P-labeled nicked DNA, either 50 µM NAD+ or no added NAD+, and LigB as specified were incubated for 15 min at 37°C. The reaction products were resolved by denaturing PAGE. The extent of ligation [36mer/(18mer + 36mer)] was determined by scanning the gel using a FUJIX phosphorimager and plotted as a function of input LigB. (B) Kinetics. Reaction mixtures containing (per 20 µl) 50 mM Tris–HCl (pH 7.5), 10 mM (NH4)2SO4, 5 mM DTT, 5 mM MgCl2, 1 pmol of 32P-labeled nicked DNA, either 50 µM NAD+ or no added NAD+, and 8 pmol of LigB were incubated at 37°C. The reactions were initiated by the addition of LigB. Aliquots (20 µl) were withdrawn at 0.25, 0.5, 1, 2, 5 and 10 min and quenched immediately with EDTA and formamide. The extent of ligation is plotted as a function of time.
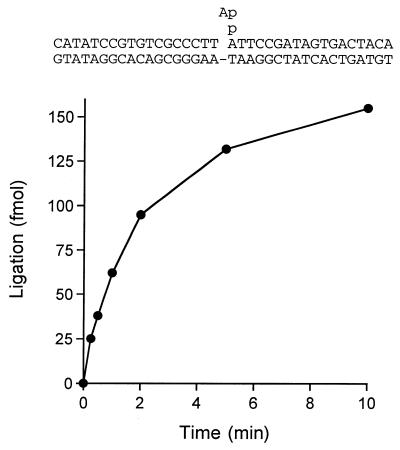
The kinetics of nick joining were examined under conditions of LigB excess (400 nM LigB and 50 nM nicked DNA). The reaction rates and extents were virtually identical with or without added NAD+ (Fig. 3B). The reaction without NAD+ reflects a single turnover of nick joining by pre-formed LigB-AMP intermediate (present at ∼56 nM concentration in the reactions). An endpoint was attained in 2–5 min at which time 80% of the input DNA substrate was sealed. We calculated an apparent rate constant of 0.026 s–1. LigB also catalyzed phosphodiester bond formation on a pre-adenylated DNA substrate (Fig. 4). Under conditions of enzyme excess (100 nM LigB and 12.5 nM nicked DNA-adenylate) LigB sealed 65% of the input DNA-adenylate in 10 min. The apparent rate constant for sealing of nicked DNA-adenylate was 0.008 s–1. The step 1 reaction of LigB with 1 µM [32P-AMP]NAD+ displayed pseudo-first order kinetics with an apparent rate constant of 0.022 s–1 (not shown).
Figure 4.
Reaction of LigB with nicked DNA-adenylate. A reaction mixture containing (per 20 µl) 50 mM Tris–HCl (pH 7.5), 10 mM (NH4)2SO4, 5 mM DTT, 5 mM MgCl2, 250 fmol of 32P-labeled nicked DNA-adenylate and 2 pmol of LigB was incubated at 37°C. The reaction was initiated by the addition of LigB. Aliquots (20 µl) were withdrawn at the times specified and quenched immediately. The extent of ligation is plotted as a function of time. The nicked DNA-adenylate substrate used in the ligation reaction is illustrated at the top.
DISCUSSION
We have demonstrated that E.coli encodes two NAD+-dependent DNA ligases. LigB lacks the tetracysteine Zn finger and the BRCT structural domains that are present in LigA and all other bacterial NAD+ ligases. In this respect LigB resembles the recently identified NAD+-dependent ligase of Amsacta moorei entomopoxvirus, a eukaryotic poxvirus (22). The AmEPV ligase, its homolog from Melanoplus sanguinipes EPV, and E.coli LigB appear to comprise a new branch of the NAD+ ligase family.
A direct comparison of the biochemical properties of purified recombinant versions of the two E.coli DNA ligases using the same nicked DNA substrate and similar reaction conditions (19, the present study, and data not shown) indicates that LigA is much more active than LigB. The specific activity of LigB under steady-state conditions is only 1% that of LigA. The activity difference is attributable to the differential response to the NAD+ cofactor. Whereas LigA is stimulated ≥100-fold by NAD+ under steady-state conditions, we find that LigB is stimulated only 3-fold.
The rate of single-turnover nick joining by LigB-adenylate (0.026 s–1) is much slower than that of LigA-adenylate (≥0.22 s–1). Thus, it appears that LigB is impaired in one or more of the component steps of the ligase pathway subsequent to adenylation of the enzyme, either in the binding of ligase-adenylate to the nicked DNA, transfer of AMP from LigB to the 5′ PO4 at the nick (step 2 chemistry), attack of the 3′ OH on the DNA-adenylate strand to form a phosphodiester (step 3 chemistry), or release of the AMP product. As we did not detect accumulation of DNA-adenylate during the single turnover reaction of LigB with nicked DNA, we presume that step 3 chemistry is not rate limiting. The apparent rate constants for step 3 under single turnover conditions were fairly similar for LigA (0.023 s–1) and LigB (0.008 s–1). As we have discussed previously (23), the isolated step 3 reaction can be paradoxically slower than the overall ligase reaction (this is the case for LigA and LigB) and we have invoked rate-limiting conformational steps in the binding of nicked DNA-adenylate to the ligase active site that do not apply during the standard nick joining reaction. The rate of the ligase-adenylation for LigB (kobs 0.022 s–1) was much slower than that for LigA (kobs ≥0.4 s–1).
Our comparison of LigB and LigA suggests that LigB is less active than LigA at multiple stages of the nick joining pathway. Moreover, we find that whereas LigA can functionally substitute for the essential Cdc9 ligase in yeast, LigB cannot do so (19 and data not shown). The simple interpretation is that the intrinsic activity of LigB activity is too weak to seal the Okazaki fragments generated during DNA replication. Can the functional differences between LigA and LigB be attributed to the structural domains that are missing from LigB? In an attempt to answer this question, we performed a domain swap in which the entire C-terminal OB-fold, zinc finger, HhH and BRCT portion of LigA (residues 315–671) was fused to the N-terminal domain Ia/nucleotidyl transferase portion of LigB (residues 1–304). However, the LigB–LigA chimera was no more active in nick joining than LigB (not shown). A second domain swap in which just the BRCT domain of LigA (residues 586–671) was fused to the C-terminus of LigB also failed to elicit a gain-of-function compared with LigB (not shown). Thus, we are unable by this strategy to engineer a more active version of LigB.
It is conceivable that LigB activity in nick joining is stimulated by other bacterial proteins, just as eukaryotic DNA ligase IV is stimulated by its binding partner XRCC4 (24). It is also possible that the function of LigB activity is not geared toward the sealing of nicks in B-form duplex DNA. It may have more specialized nucleic acid substrate preferences. A genetic analysis of the effects of ligB deletion on DNA transactions in E.coli may be illuminating in this regard.
Finally, it is worth noting that LigB is not restricted to E.coli. The genome of Yersinia pestis, the causative agent of plague, appears to encode two NAD+-dependent ligases (25). One Yersinia ligase is a 670 amino acid homolog of E.coli LigA and the second is a 562 amino acid polypeptide with 43% side-chain identity to E.coli LigB. Just as in E.coli, the Yersinia ligB gene is located adjacent to the gene for guanylate kinase. As more and more bacterial genomes are sequenced, it will be of interest to see how many species other than E.coli and Yersinia have multiple NAD+-dependent ligases.
Acknowledgments
ACKNOWLEDGEMENTS
Supported by NIH grant GM63611.
REFERENCES
- 1.Gellert M. (1967) Formation of covalent circles of lambda DNA by E.coli extracts. Proc. Natl Acad. Sci. USA, 57, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman S.B., Little,J.W., Oshinky,C.K. and Gellert,M. (1967) Enzymatic joining of DNA strands: a novel reaction of diphosphopyridine nucleotide. Proc. Natl Acad. Sci. USA, 57, 1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivera B.M. and Lehman,I.R. (1967) Diphosphopyridine nucleotide: a cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc. Natl Acad. Sci. USA, 57, 1700–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivera B.M. and Lehman,I.R. (1967) Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc. Natl Acad. Sci. USA, 57, 1426–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehman I.R. (1974) DNA ligase: structure, mechanism, and function. Science, 186, 790–797. [DOI] [PubMed] [Google Scholar]
- 6.Singleton M.R., Håkansson,K., Timson,D.J. and Wigley,D.B. (1999) Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure, 7, 35–42. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.Y., Chang,C., Song,H.K., Moon,J., Yang,J., Kim,H.K., Kwon,S.T. and Suh,S.W. (2000) Crystal structure of NAD+-dependent DNA ligase: modular architecture and functional implications. EMBO J., 19, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuman S. and Schwer,B. (1995) RNA capping enzyme and DNA ligase—a superfamily of covalent nucleotidyl transferases. Mol. Microbiol., 17, 405–410. [DOI] [PubMed] [Google Scholar]
- 9.Subramanya H.S., Doherty,A.J., Ashford,S.R. and Wigley,D.B. (1996) Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]
- 10.Odell M., Sriskanda,V., Shuman,S. and Nikolov,D. (2000) Crystal structure of eukaryotic DNA ligase–adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell, 6, 1183–1193. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman M.M., Hicks,M.L. and Gellert,M. (1973) Genetics and function of DNA ligase in Escherichia coli. J. Mol. Biol., 77, 531–547. [DOI] [PubMed] [Google Scholar]
- 12.Konrad E.B., Modrich,P. and Lehman,I.R. (1973) Genetic and enzymatic characterization of a conditional lethal mutant of Escherichia coli K12 with a temperature-sensitive DNA ligase. J. Mol. Biol., 77, 519–529. [DOI] [PubMed] [Google Scholar]
- 13.Park U.E., Olivera,B.M., Hughes,K.T., Roth,J.R. and Hillyard,D.R. (1989) DNA ligase and the pyridine cycle in Salmonella typhimurium. J. Bacteriol., 171, 2173–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petit M.A. and Ehrlich,S.D. (2000) The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res., 28, 4642–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaczmarek F.S., Zaniewski,R.P., Gootz,T.D., Danley,D.E., Mansour,M.N., Griffor,M., Kamath,A.V., Cronan,M., Mueller,J., Sun,D. et al. (2001) Cloning and functional characterization of an NAD+-dependent DNA ligase from Staphylococcus aureus. J. Bacteriol., 183, 3016–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng C. and Shuman,S. (1997) Characterization of an ATP-dependent DNA ligase encoded by Haemophilus influenzae. Nucleic Acids Res., 25, 1369–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson A., Day,J. and Bowater,R. (2001) Bacterial DNA ligases. Mol. Microbiol., 40, 1241–1248. [DOI] [PubMed] [Google Scholar]
- 18.Luo J. and Barany,F. (1996) Identification of essential residues in Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sriskanda V., Schwer,B., Ho,C.K. and Shuman,S. (1999) Mutational analysis of E.coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4.Nucleic Acids Res., 27, 3953–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doherty A.J. and Suh,S.W. (2000) Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res., 28, 4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 22.Sriskanda V., Moyer,R.W. and Shuman,S. (2001) NAD+-dependent DNA ligase encoded by a eukaryotic virus. J. Biol. Chem., 276, 36100–36109. [DOI] [PubMed] [Google Scholar]
- 23.Sriskanda V. and Shuman,S. (1988) Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res., 26, 4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill J., Wren,B.W., Thomson,N.R., Titball,R.W., Holden,M.T.G., Prentice,M.B., Sebaihia,M., James,K.D., Churcher,C., Mungall,K.L. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]