Figure 9.
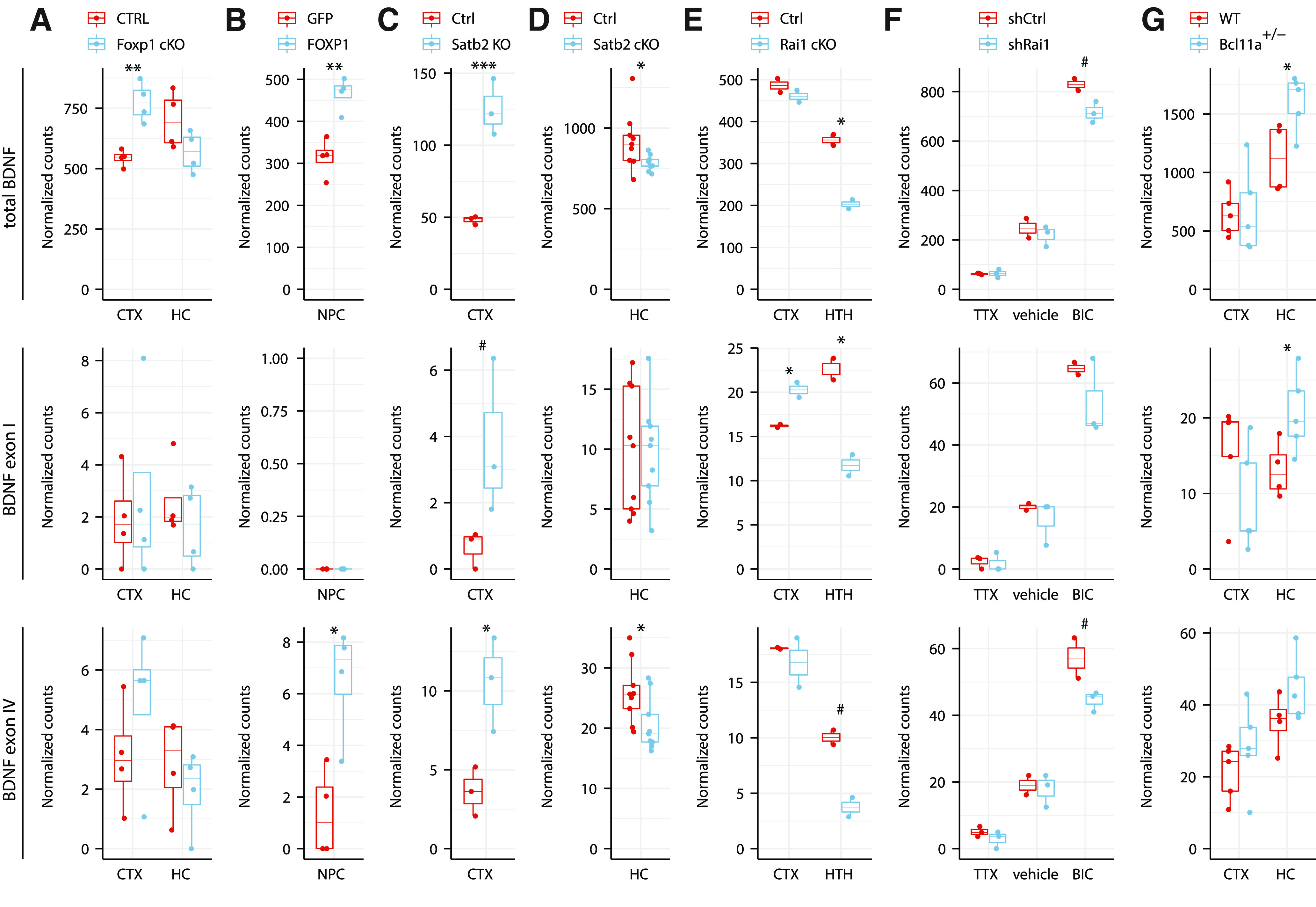
Transcription factors FOXP1, SATB2, RAI1, BCL11A regulate BDNF expression in a brain region-specific manner. Total BDNF levels (upper panels), levels of BDNF exon I (middle panels), and BDNF exon IV (lower panels) were analyzed from previously published RNA-seq experiments: (A) postnatal day 47 adult male mice cortex (CTX) and hippocampus (HC) in pyramidal neuron-specific Foxp1 conditional knock-out (cKO) animals using Emx1-Cre driver (Araujo et al., 2017); (B) human neural progenitor cells (NPC) overexpressing GFP or FOXP1 (Araujo et al., 2015); (C) postnatal day 0 CTX of Satb2 knock-out animals (McKenna et al., 2015); (D) adult mice HC CA1 region in Satb2 conditional knock-out in adult neurons using Camk2a-Cre driver (Jaitner et al., 2016); (E) three-week-old Rai1 flox/flox (Ctrl) and Rai1 cKO using Nestin-Cre driver in CTX and eight-week-old Rai1 cKO using Vglut2-Cre driver in hypothalamus (HTH; W.H. Huang et al., 2016); (F) Bru-seq to determine nascent transcripts in cultured cortical and hippocampal neurons derived from embryonic day 18 mice, grown 17 DIV and infected with lentiviruses expressing shRNA against Rai1 (shRai1) and treated with either tetrodotoxin (TTX), vehicle, or bicuculline (BIC) for 4 h (Garay et al., 2020); (G) CTX and HC of 16-week-old male wild-type (WT) or Bcl11a heterozygous knock-out mice (Dias et al., 2016). RNA-seq counts were normalized with DESeq2 and are shown as box plots, where the hinges show 25% and 75% quartiles, the horizontal line shows the median value, the upper whisker extends from the hinge to the largest value no further than 1.5× interquartile range from the hinge, the lower whisker extends from the hinge to the smallest value at most 1.5× interquartile range of the hinge. All data points are shown with dots. Statistical analysis was performed on log-transformed data. #p < 0.1, *p < 0.05, **p < 0.01, ***p < 0.001, unpaired equal variance two tailed t test.
