Figure 5: Combining embryo single-cell analysis and neoblast cell-lineage tracing reveals a molecular trajectory for neoblasts during development.
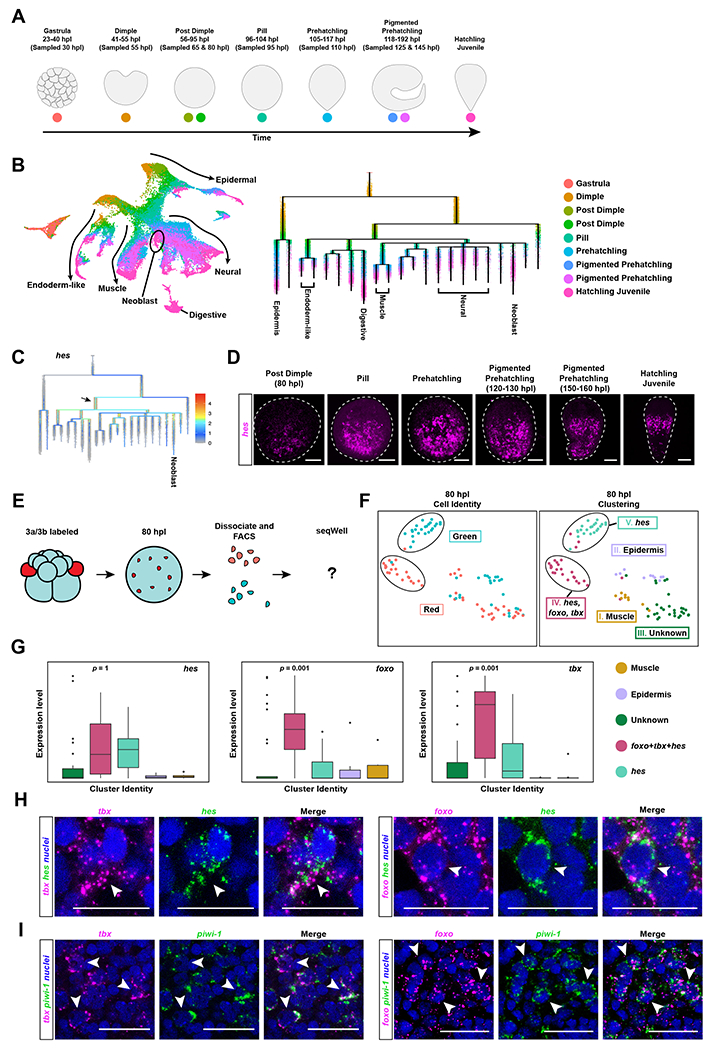
A) Schematic highlighting Hofstenia developmental stages and the developmental timing at which they were sampled to generate the single-cell RNA-seq dataset. (B) Left: UMAP plot of the embryonic dataset merged with the Hatchling Juvenile dataset16 to obtain endpoints in development. Arrows and tissue-type labels highlight the UMAP topology, and the expression of known marker genes of the corresponding tissues as shown in Fig. S5A. Right: URD trajectory inference tree showing the putative molecular trajectories of cells in the form of a tree, with the branch tips annotated with known differentiated cell types16. Dots, representing cells, are colored based on developmental time points shown in A. (C) Expression levels of the gene hes projected onto the URD tree. We see high expression levels of hes at an internode that is populated largely by cells from the Post Dimple stage (80 hours post laying (hpl)) (black arrow) (D) in situ hybridization of hes reveals expression first detectable at the Post Dimple stage (80 hpl) which culminated in a neoblast-like pattern in hatchlings. (E) Schematic workflow for sorting and sequencing a total of 96 cells from Post Dimple stage (80 hpl) embryos with photoconverted 3a/3b progeny at single-cell resolution. Red cells represent 3a/3b progeny that should make neoblasts, whereas green cells are progeny of other blastomeres that give rise to differentiated cell types. (F) Left: UMAP showing photoconversion status (red, 3a/3b progeny; cyan, progeny of other, unconverted blastomeres). Right: UMAP showing cluster identities. Muscle (Cluster I; mustard) and epidermal (Cluster II; light purple) cells were detected. One cluster did not yield any markers with enriched expression, and is labeled as “Unknown” (Cluster III; dark green). Clusters that were predominantly composed of 3a/3b progeny (Cluster IV; cranberry) and predominantly composed unconverted progeny (Cluster V; light green) were detected (circled in black). Both of these clusters expressed hes, but the Cluster IV, composed of 3a/3b progeny, show significantly enriched expression of foxo and tbx. (G) Box plots showing hes was expressed in both of Cluster IV and Cluster V, corroborating the lineage analysis in B that hes marks undifferentiated embryonic progenitors for many cell types, foxo Iand tbx were highly enriched in Cluster V, which consists primarily of 3a/3b progeny. Data represented as mean ± SEM. (H) Double in situ hybridization of foxo and tbx with hes in Pigmented Prehatchling (120 hpl) embryos shows co-localization (white arrowheads). (I) Double in situ hybridization of foxo and tbx with piwi-1 in hatched worms shows co-localization (white arrowheads), confirming that both foxo and tbx mark a subset of neoblasts. Scale bars for embryo and hatchling images = 100μm. Scale bars for high magnification images = 50μm. See also Figure S5, Table S1, Table S2, and Table S3.
