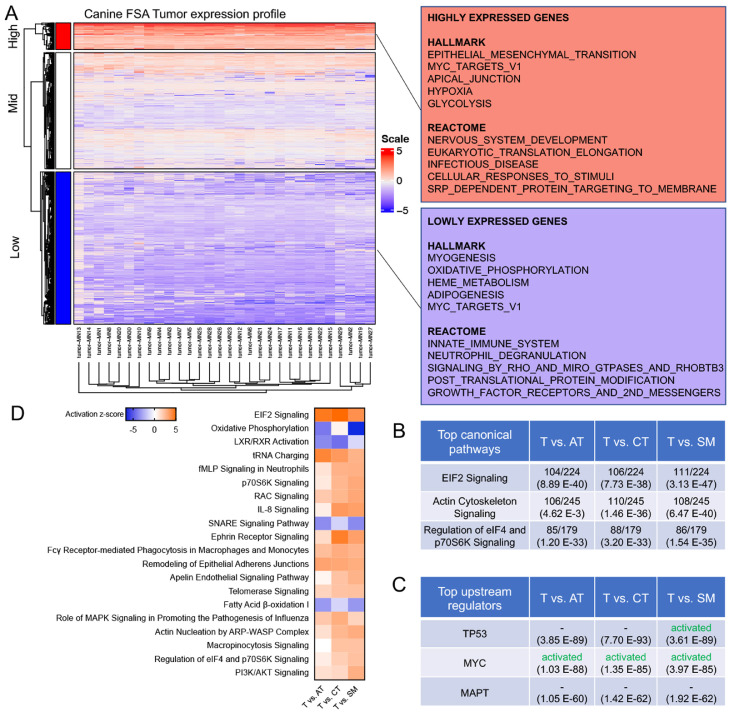
Fig. 4.
Molecular features of canine FSA. A) (left) Heatmap of all cases using all proteins showing highly homogeneous protein expression across all FSA cases. (right) GSEA using HALLMARK and REACTOME pathway databases of highly (top) and lowly (bottom) expressed proteins in FSA. B) Top canonical pathways detected in T vs AT, CT and SM using Ingenuity pathway analysis (IPA). Values on the top of a cell indicate how many targets were detected in the dataset/how many targets are attributed to the respective dataset, and values in brackets represent p-values. C) Top upstream regulators detected in T vs AT, CT and SM using Ingenuity pathway analysis (IPA). The top half of a cell indicates the predicted activation status, and values in brackets represent p-values. D) Activation status of top 20 canonical pathways differentially regulated between T vs AT, CT and SM.