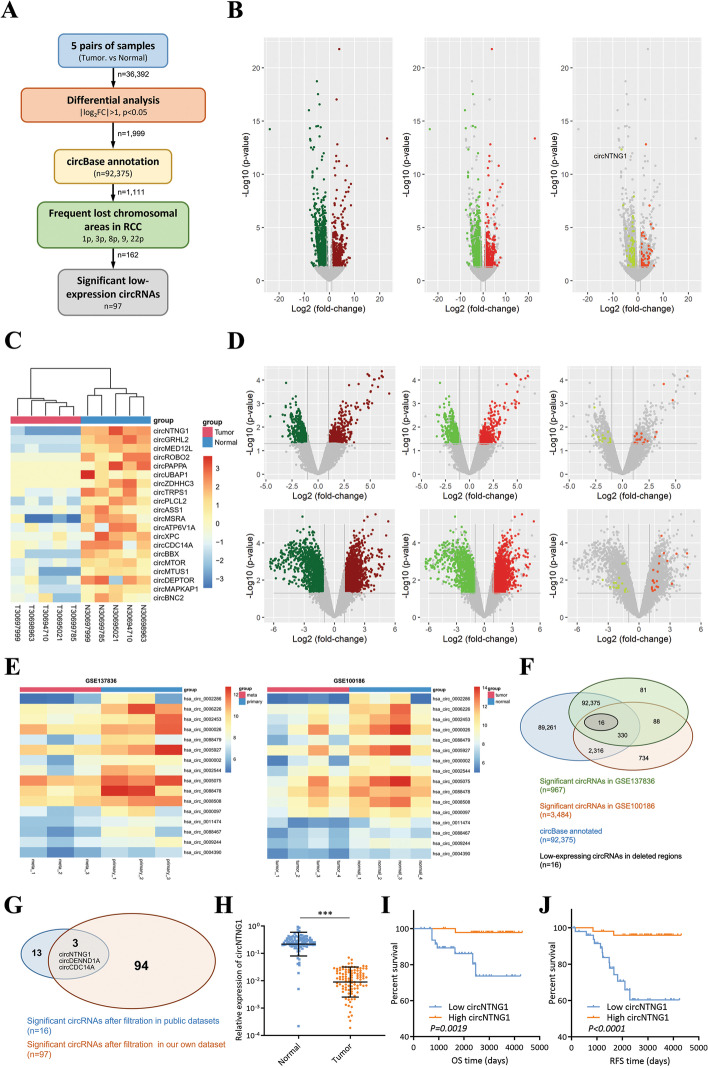
Fig. 1.
Sequencing and microarray datasets reveal tumor-suppressing circNTNG1 in RCC. a. Workflow of filtering frequently deleted circRNAs in RCC using our own patient dataset. Numbers near the arrows in each steps indicated the remaining circRNAs. b. Volcano plots of the remaining circRNAs during each step. Our own dataset was used. Colored dots indicated significant circRNAs. Grey dots indicated circRNAs that were filtered out. c. Heatmap of the top 20 frequently deleted circRNAs in our own circRNA sequencing dataset. d. Volcano plots of the remaining circRNAs during each step. Two public RCC circRNA microarray datasets were used. Colored dots indicated significant circRNAs. Grey dots indicated circRNAs that were filtered out. e. Heatmaps of the 16 frequently deleted circRNAs in two public RCC circRNA microarray datasets. f. Venn gram of the frequent deleted circRNAs selection in two public RCC circRNA microarray datasets. g. Venn gram of intersection of the two public RCC circRNA datasets and our own circRNA sequencing dataset. h. Expression analysis of circNTNG1 in tumors and normal adjacent tissues in our patient cohort (total 102 patients). i. Survival analysis (overall survival) of circNTNG1 in our patient cohort. j. Survival analysis (recurrent-free survival) of circNTNG1 in our patient cohort. Data are mean ± SD, n = 3