Fig. 3.
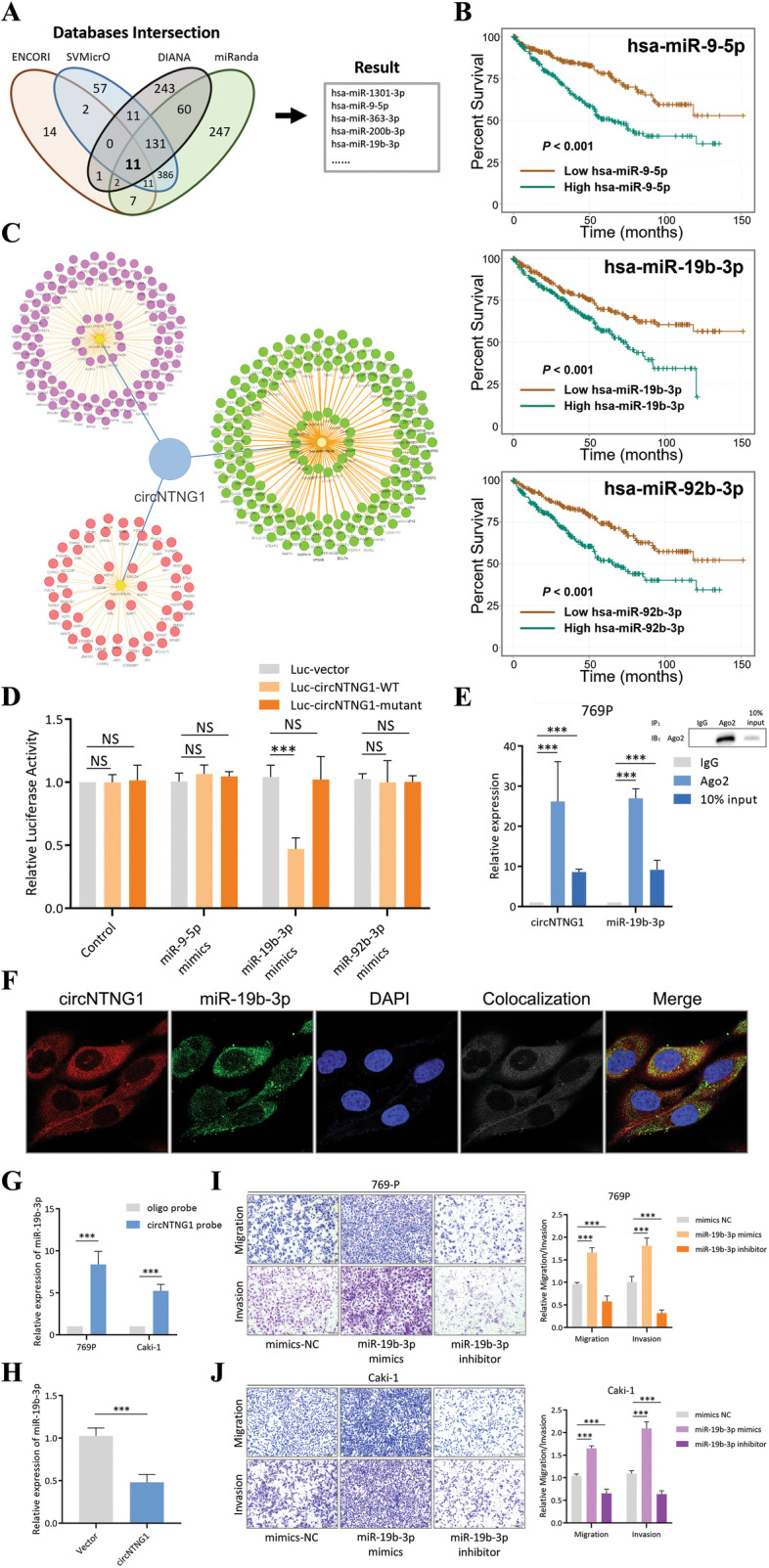
CircNTNG1 exerted a sponge effect on miR-19b-3p. a. Venn gram of finding the potential interacting miRNAs of a circNTNG1. The final miRNAs were generated by intersecting the results of four different online databases (SVMicrO, DIANA-microT, ENCORI, and miRanda). b. Survival analysis of three potential miRNAs involved in the circNTNG1 regulatory axis on TCGA KIRC patients. c. The circNTNG1/miRNA/mRNA interaction network. d. Luciferase reporter assay with vector, circNTNG1 wild-type sequence and circNTNG1 mutant sequence transfected with three different miRNA mimics. Vector group was used as normalization control. e. RNA immunoprecipitation using an Ago2 antibody and qRT-PCR quantification of circNTNG1 and miR-19b-3p in the pull-down product. IgG group was used as normalization control. f. FISH co-localization experiment of circNTNG1 and miR-19b-3p. DAPI was used to indicate the location of nuclei. g. RNA pull-down experiment with a circNTNG1 specific probe. An oligo probe was used a negative control. h. qRT-PCR quantification of miR-19b-3p in 769P cells transfected with control vector/circNTNG1 overexpression. The control vector group was used as normalization. i. Representative images (left) and quantification (right) data of Transwell migration/invasion assay of 769P cells with miR-19b-3p mimics/miR-19b-3p inhibitor. j. Representative images (left) and quantification (right) data of Transwell migration/invasion assay of Caki-1 cells with miR-19b-3p mimics/miR-19b-3p inhibitor. Data are mean ± SD, n = 3
