Figure 1.
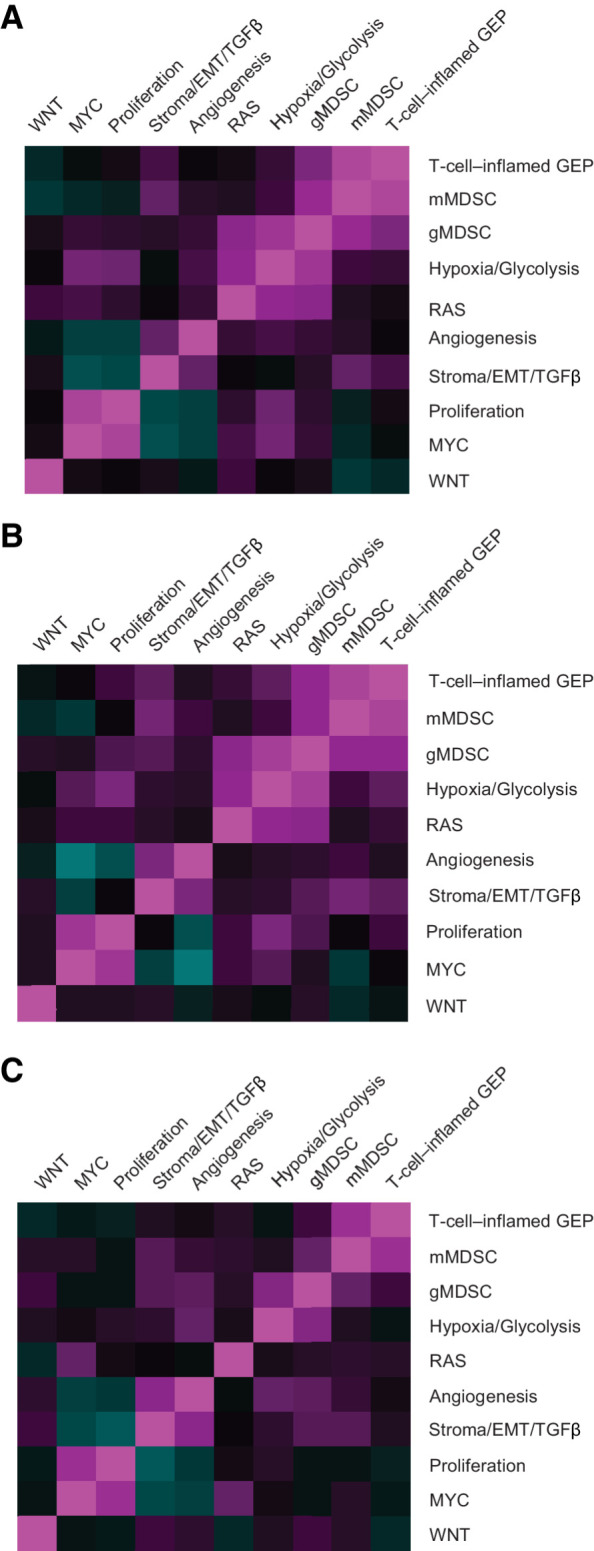
Pairwise Spearman correlation of consensus signatures. Merck–Moffitt dataset (A), TCGA dataset (B), and pooled pembrolizumab trial analysis population (C). EMT, epithelial-to-mesenchymal transition; GEP, gene expression profile; gMDSC, granulocytic myeloid-derived suppressor cell; mMDSC, monocytic myeloid-derived suppressor cell; TCGA, The Cancer Genome Atlas; TGFβ, transforming growth factor β.
