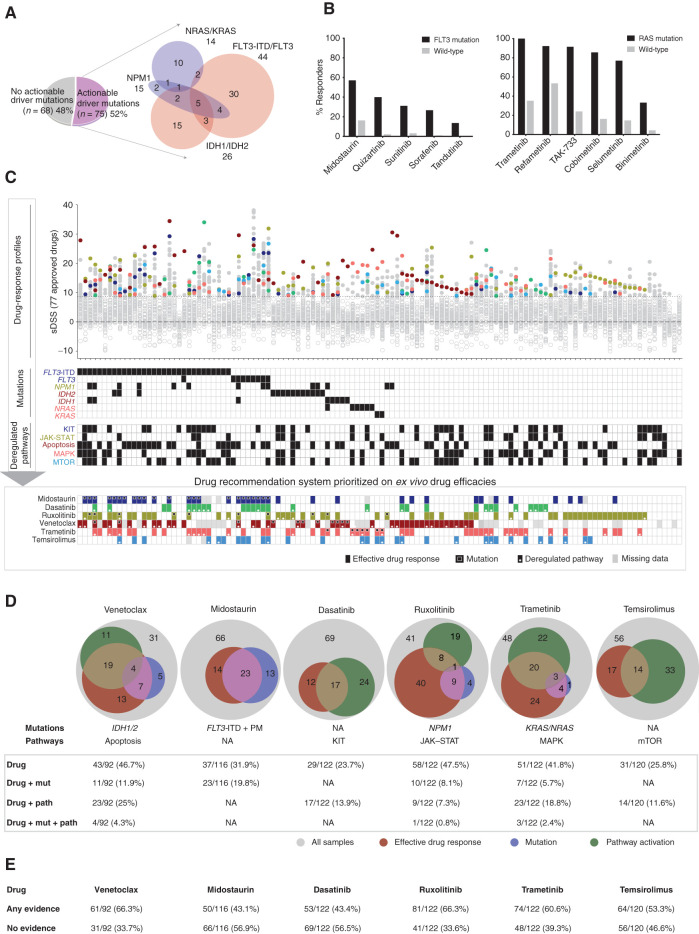
Figure 5.
Genomic and transcriptomics-based prediction of ex vivo drug efficacies. A, The division of 143 AML patient samples in actionable and nonactionable subsets. B,Ex vivo drug sensitivity of FLT3i in FLT3-ITD and point mutation–positive samples and of MEKi in KRAS/NRAS mutation–positive samples. C, Samples with complete molecular profiling and drug-response data ordered as per actionable driver mutations and subsequently nonactionable mutations. Selective drug responses for FDA/EMA-approved 77 drugs are depicted on the Y-axis and individual patient samples on the X-axis, where ineffective drugs below sDSS 8.7 were marked with gray rings. The common effective drugs were highlighted for integration with mutation and pathway activation. The bottom panel illustrates integrated ex vivo efficacy and the presence of respective mutations and pathways for each sample. D, Statistics of patient samples showing evidence of drug sensitivity, the presence of mutation, and pathway activation for key targeted drugs in AML including the BCL2i venetoclax, FLT3i midostaurin, TKi dasatinib, JAKi ruxolitinib, MEKi trametinib, and JAKi ruxolitinib. E, The drug-wise percentage of samples showing any evidence and no evidence from effective drug response, mutation, and/or pathway upregulation.