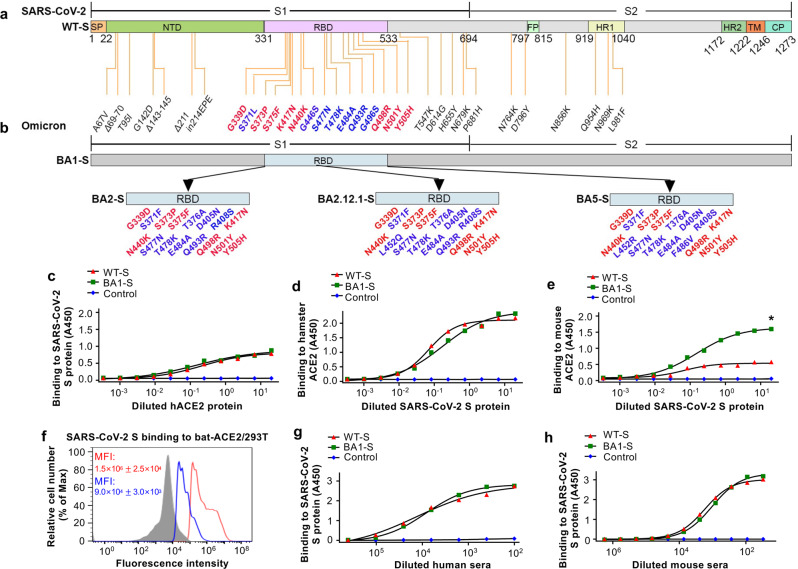
Fig. 1. Schematic map of spike (S) protein of SARS-CoV-2 original strain and Omicron subvariants and characterization of SARS-CoV-2 BA1-S protein.
Schematic map of SARS-CoV-2 S protein of the original wild-type (WT-S) strain (a) and Omicron BA1 subvariant (BA1-S) (b). Mutant amino acid residues of Omicron BA1 subvariant are shown in the S1 (including N-terminal domain (NTD) and receptor-binding domain (RBD)) and S2 subunits of S protein, respectively. Mutant amino acid residues in the RBD of Omicron BA2, BA2.12.1, and BA5 are shown (b). SP signal peptide. FP fusion peptide. HR1 and HR2, heptad repeat regions 1 and 2. TM transmembrane domain. CP cytoplasmic tail. ELISA analysis of binding of Omicron BA1-S protein (BA1-S) or original S protein (WT-S) to human angiotensin-converting enzyme 2 (hACE2) (c), hamster ACE2 (d), and mouse ACE2 (e) proteins, respectively. Control, PBS. Statistical significance between the binding of WT-S and BA1-S proteins to mouse ACE2 protein was analyzed using a two-tailed student t-test, and * (P < 0.05) indicates a significant difference. f Flow cytometry analysis of binding of BA1-S (blue line) and WT-S (red line) proteins to bat ACE2-expressing 293T cells (bat-ACE2/293T). 293T cells were transiently transfected with bat ACE2 plasmid and incubated with each protein (5 µg/ml) for analysis by flow cytometry. Gray shading, mock-incubated cells. MFI, median fluorescence intensity. ELISA for detection of binding of WT-S and BA1-S proteins to SARS-CoV-2 S-vaccinated human (g) and mouse (h) serum-neutralizing antibodies, respectively. The data (in c–h) are expressed as mean ± standard deviation of the mean (s.e.m.) of the duplicate to quadruple wells. The experiments were repeated twice, leading to similar results.