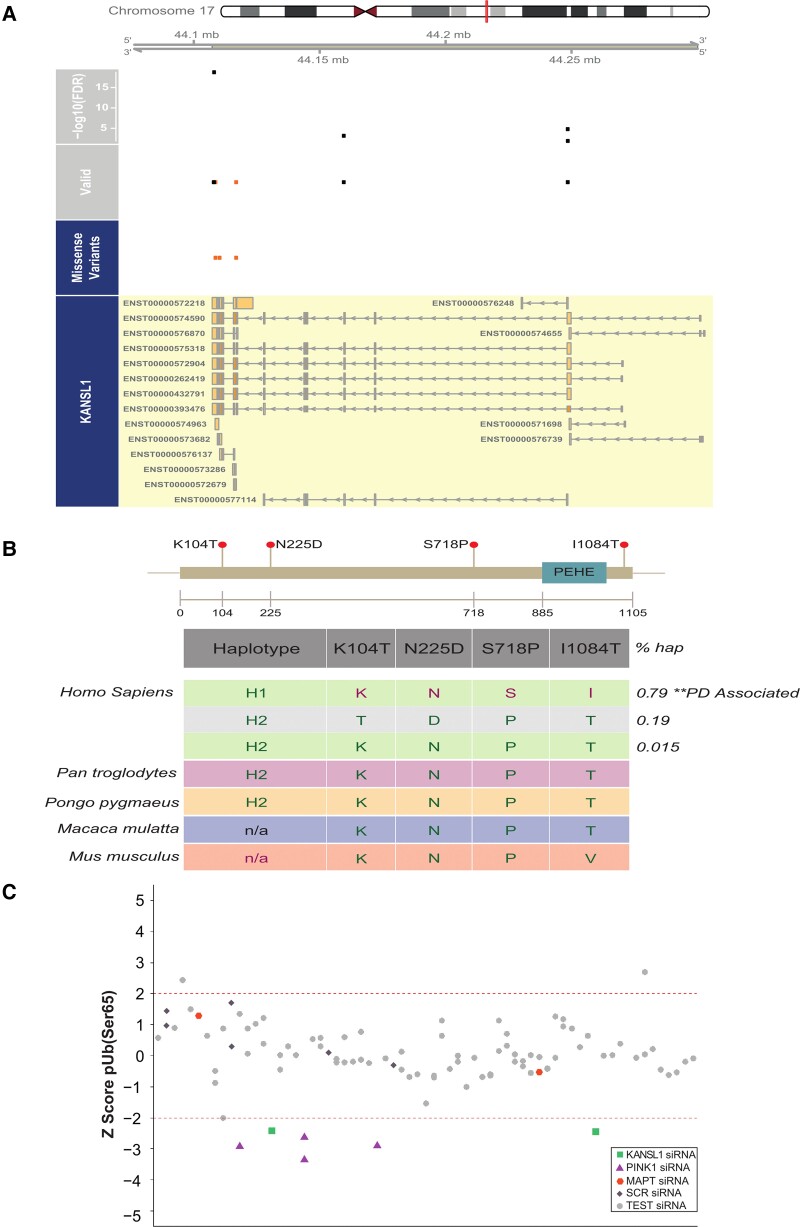
Figure 6.
KANSL1 presents ASE sites in LD with the H1/H2 SNP, and pUb(Ser65) levels are altered by siRNA KD of KANSL1 but not other genes present at the 17q21 locus. (A) ASEs derived from putamen and substantia nigra in high LD with the H1/H2 tagging SNP, rs12185268 and their position along the KANSL1 gene. The missense variants track displays the variants annotated as missense by gnomAD v.2.1.1.70 The valid track displays the heterozygous sites (orange = missense) with an average read depth >15 reads across all samples, which were examined for ASE. The topmost track displays the false discovery rate-corrected minimum −log10P-value across samples for the sites that show an ASE in at least one sample. (B) Conservation of the KANSL1 protein across species. The four coding variants (NM_001193465) in the KANSL1 gene are in high LD (r2 > 0.8) with the H1/H2 haplotypes. (C) pUb(Ser65) Z scores of one representative 17q21 locus screen plate. See Supplementary Table 10 for the complete list of the genes screened.