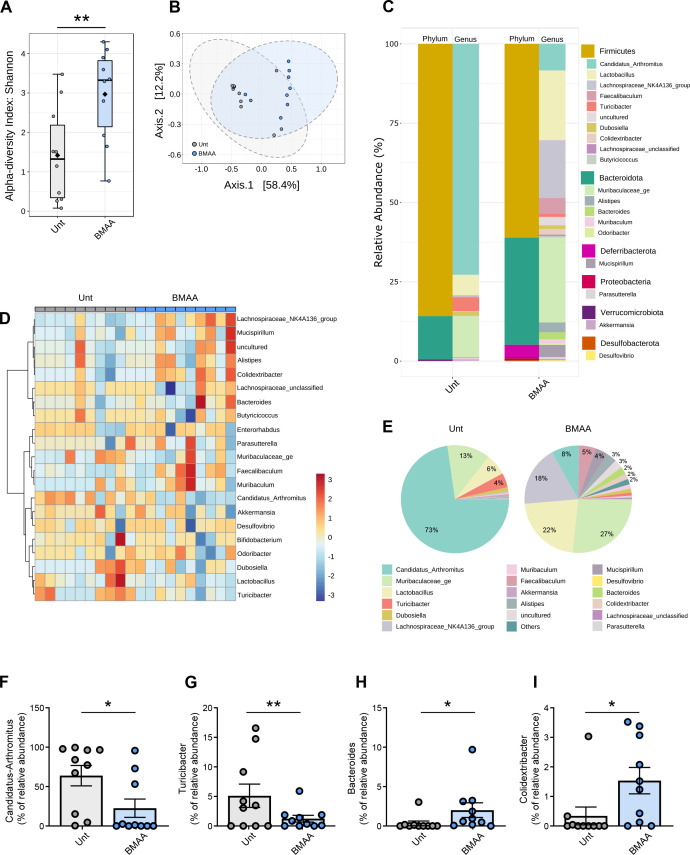
Figure 1.
Ileum mucosa-associated microbiome diversity in β-N-methylamino-L-alanine (BMAA)-treated mice. (A) Alpha diversity measured using the Shannon index at operational taxonomic unit (OTU) level derived from 16S rDNA sequencing of ileum intestinal samples from untreated (Unt) or BMAA-treated mice (n values for Unt=10 and BMAA=10, Unt vs BMAA, Mann-Whitney test, **p=0.0089). (B) Beta diversity evaluated by principal coordinate analysis (PCoA) based on Bray-Curtis index of OTUs derived from 16S rDNA sequencing of ileum intestinal samples from Unt or BMAA-treated mice (n values for Unt=10 and BMAA=10; PERMANOVA: r2=0.205, *p<0.015; PERMDISP: F=0.773, p=0.391). (C) Taxonomic diversity of ileum intestinal samples from Unt or BMAA-treated mice at the phylum and genus levels. (D) Heatmap of the relative abundances of genera detected in ileum intestinal samples from Unt or BMAA-treated mice using Pearson’s correlation coefficient as a distance metric, with clustering based on Ward’s algorithm. (E) Pie-charts showing the proportional taxonomic composition at the genus level of ileum intestinal microbiota samples from Unt or BMAA-treated mice. (F–I) Differential abundance of selected genera in ileum intestinal samples from Unt or BMAA-treated mice (n values for Unt=10 and BMAA=10, Unt vs BMAA, DESeq2 statistical analysis). (F) ‘Candidatus Arthromitus’ (*padj=0.0296). (G) Turicibacter (**padj=0.0027). (H) Bacteroides (*padj=0.0150). (I) Colidextribacter (*padj=0.0103).